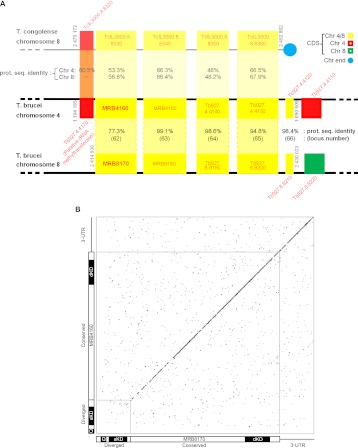
FIGURE 1.
The paralogous MRB8170 and MRB4160 genes. (A) Scheme showing that the kMAP genes are part of an array of five paralogs retained on both chromosomes 4 and 8 arising from a duplication event (bottom two black lines). In addition, a syntenic region of T. congolense chromosome 8 (top gray line) is aligned to the gene array present on T. brucei chromosome 4. CDSs are depicted as boxes with either its name or locus tag given inside or adjacently just above or below the box in red letters. MRB8170 and MRB4160 loci are in bold. Paralogs retained on both T. brucei chromosome 4 and 8 are yellow, only on the former in red and the latter in green. The top number in the shaded region between the duplicated CDSs is the percentage protein sequence identity between them according to a ClustalW alignment performed in the BioEdit program. The bottom number in parentheses is the locus number as given by Jackson (2007), from which this figure is adapted. The numbers in the shaded region between T. congolense chromosome 8 and T. brucei chromosome 4 represent the protein sequence identity of the CDSs from the former with T. brucei chromosomes 8 (top) and 4 (bottom) as calculated previously. The chromosome location at the beginning and end of the array of genes in question is given in gray adjacent to the relevant bounding CDS. (B) Dot-plot comparing the CDSs plus 3′-UTRs of MRB8170 (x-axis) and MRB4160 (y-axis) to reveal regions of nucleotide sequence identity. Nucleotide matches are represented as a black dot. The diagonal represents the identical regions of the two sequences. The small gray square in the lower left-hand corner encloses the diverged region of two sequences. Schemes depicting the relevant regions of MRB8170 and MRB4160 are provided along the axes. The diverged and conserved regions are indicated, while the 3′-UTR is shown as a black line. The regions targeted for the single (sKD) and double (dKD) knockdowns and regions amplified by quantitative real-time PCR primers (Q) are shown.