Abstract
We have synthesized two oligodeoxyribonucleotide mixtures that contain sequences complementary to different parts of the hypothetical mRNA sequence of xenopsin, a biologically active octapeptide found in skin extracts from Xenopus laevis. The two primer pools were independently used to initiate reverse transcription on skin poly(A)+ RNA and the resulting cDNAs were then used to screen in parallel a cDNA library prepared from skin poly(A)+ RNA. One of the clones that hybridized with both probes was subjected to sequence analysis. It contains a nearly full-length DNA copy of a mRNA of approximately equal to 490 nucleotides that encodes a xenopsin precursor protein. The deduced precursor is 80 amino acids long, exhibits a putative signal sequence at the NH2 terminus, and contains the biologically active peptide at the COOH terminus. The region corresponding to the NH2-terminal portion of the xenopsin precursor shows a striking nucleotide and amino acid sequence homology with the precursor of PYLa, another recently described peptide from Xenopus skin.
Full text
PDF

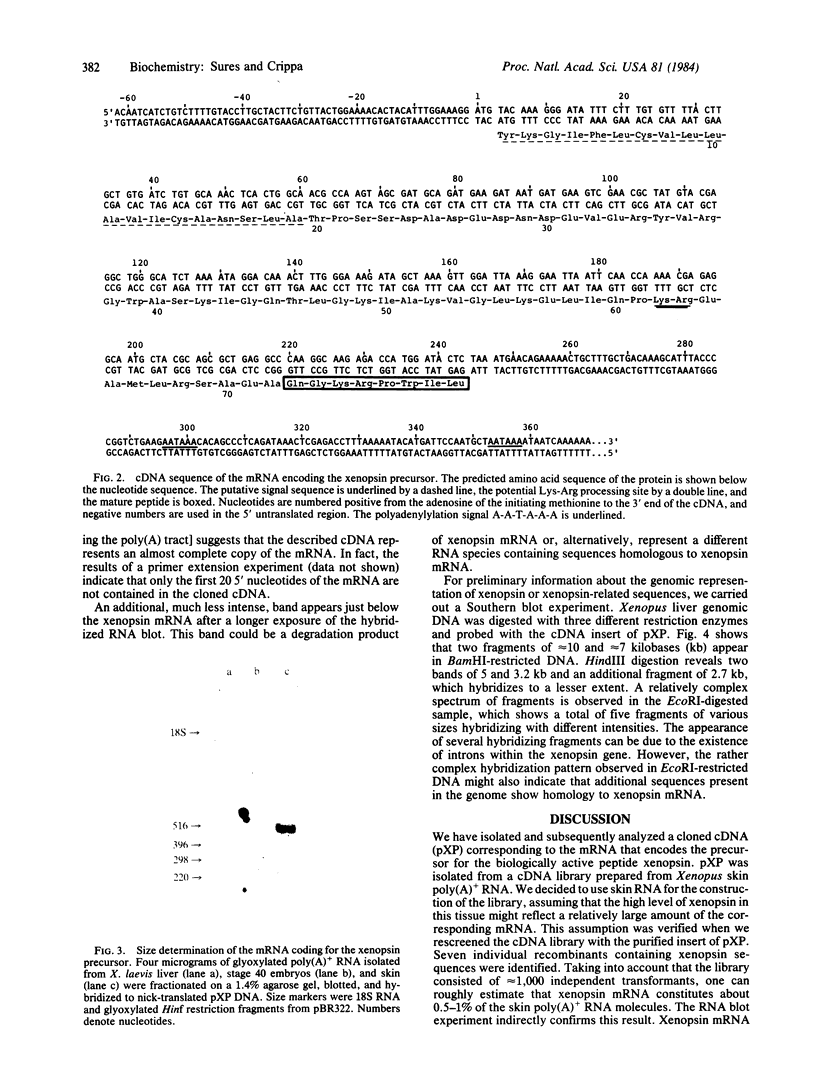


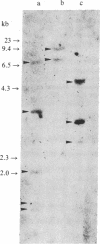
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Araki K., Tachibana S., Uchiyama M., Nakajima T., Yasuhara T. Isolation and structure of a new active peptide "Xenopsin" on the smooth muscle, especially on a strip of fundus from a rat stomach, from the skin of Xenopus laevis. Chem Pharm Bull (Tokyo) 1973 Dec;21(12):2801–2804. doi: 10.1248/cpb.21.2801. [DOI] [PubMed] [Google Scholar]
- Araki K., Tachibana S., Uchiyama M., Nakajima T., Yasuhara T. Isolation and structure of a new active peptide xenopsin on rat stomach strip and some biogenic amines in the skin of Xenopus laevis. Chem Pharm Bull (Tokyo) 1975 Dec;23(12):3132–3140. doi: 10.1248/cpb.23.3132. [DOI] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G., Dobberstein B. Transfer of proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J Cell Biol. 1975 Dec;67(3):852–862. doi: 10.1083/jcb.67.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraway R., Ruane S. E., Feurle G. E., Taylor S. Amphibian neurotensin (NT) is not xenopsin (XP): dual presence of NT-like and XP-like peptides in various amphibia. Endocrinology. 1982 Apr;110(4):1094–1101. doi: 10.1210/endo-110-4-1094. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Erspamer V. Biogenic amines and active polypeptides of the amphibian skin. Annu Rev Pharmacol. 1971;11:327–350. doi: 10.1146/annurev.pa.11.040171.001551. [DOI] [PubMed] [Google Scholar]
- Gubler U., Seeburg P., Hoffman B. J., Gage L. P., Udenfriend S. Molecular cloning establishes proenkephalin as precursor of enkephalin-containing peptides. Nature. 1982 Jan 21;295(5846):206–208. doi: 10.1038/295206a0. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Meselson M. Plasmid screening at high colony density. Gene. 1980 Jun;10(1):63–67. doi: 10.1016/0378-1119(80)90144-4. [DOI] [PubMed] [Google Scholar]
- Hoffmann W., Bach T. C., Seliger H., Kreil G. Biosynthesis of caerulein in the skin of Xenopus laevis: partial sequences of precursors as deduced from cDNA clones. EMBO J. 1983;2(1):111–114. doi: 10.1002/j.1460-2075.1983.tb01390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann W., Richter K., Kreil G. A novel peptide designated PYLa and its precursor as predicted from cloned mRNA of Xenopus laevis skin. EMBO J. 1983;2(5):711–714. doi: 10.1002/j.1460-2075.1983.tb01489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Ike Y., Ikuta S., Itakura K. Solid phase synthesis of polynucleotides. VI. Further studies on polystyrene copolymers for the solid support. Nucleic Acids Res. 1982 Mar 11;10(5):1755–1769. doi: 10.1093/nar/10.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius D., Blair L., Brake A., Sprague G., Thorner J. Yeast alpha factor is processed from a larger precursor polypeptide: the essential role of a membrane-bound dipeptidyl aminopeptidase. Cell. 1983 Mar;32(3):839–852. doi: 10.1016/0092-8674(83)90070-3. [DOI] [PubMed] [Google Scholar]
- Kakidani H., Furutani Y., Takahashi H., Noda M., Morimoto Y., Hirose T., Asai M., Inayama S., Nakanishi S., Numa S. Cloning and sequence analysis of cDNA for porcine beta-neo-endorphin/dynorphin precursor. Nature. 1982 Jul 15;298(5871):245–249. doi: 10.1038/298245a0. [DOI] [PubMed] [Google Scholar]
- Kreil G., Haiml L., Suchanek G. Stepwise cleavage of the pro part of promelittin by dipeptidylpeptidase IV. Evidence for a new type of precursor--product conversion. Eur J Biochem. 1980 Oct;111(1):49–58. doi: 10.1111/j.1432-1033.1980.tb06073.x. [DOI] [PubMed] [Google Scholar]
- Land H., Schütz G., Schmale H., Richter D. Nucleotide sequence of cloned cDNA encoding bovine arginine vasopressin-neurophysin II precursor. Nature. 1982 Jan 28;295(5847):299–303. doi: 10.1038/295299a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nakanishi S., Inoue A., Kita T., Nakamura M., Chang A. C., Cohen S. N., Numa S. Nucleotide sequence of cloned cDNA for bovine corticotropin-beta-lipotropin precursor. Nature. 1979 Mar 29;278(5703):423–427. doi: 10.1038/278423a0. [DOI] [PubMed] [Google Scholar]
- Noda M., Furutani Y., Takahashi H., Toyosato M., Hirose T., Inayama S., Nakanishi S., Numa S. Cloning and sequence analysis of cDNA for bovine adrenal preproenkephalin. Nature. 1982 Jan 21;295(5846):202–206. doi: 10.1038/295202a0. [DOI] [PubMed] [Google Scholar]
- Okayama H., Berg P. High-efficiency cloning of full-length cDNA. Mol Cell Biol. 1982 Feb;2(2):161–170. doi: 10.1128/mcb.2.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller R. H., Jackson J. F., McAllister L. B., Rothman B. S., Mayeri E., Axel R. A single gene encodes multiple neuropeptides mediating a stereotyped behavior. Cell. 1983 Jan;32(1):7–22. doi: 10.1016/0092-8674(83)90492-0. [DOI] [PubMed] [Google Scholar]
- Spohr G., Reith W., Sures I. Organization and sequence analysis of a cluster of repetitive DNA elements from Xenopus laevis. J Mol Biol. 1981 Oct 5;151(4):573–592. doi: 10.1016/0022-2836(81)90424-1. [DOI] [PubMed] [Google Scholar]
- Taylor W. L., Collier K. J., Deschenes R. J., Weith H. L., Dixon J. E. Sequence analysis of a cDNA coding for a pancreatic precursor to somatostatin. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6694–6698. doi: 10.1073/pnas.78.11.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]