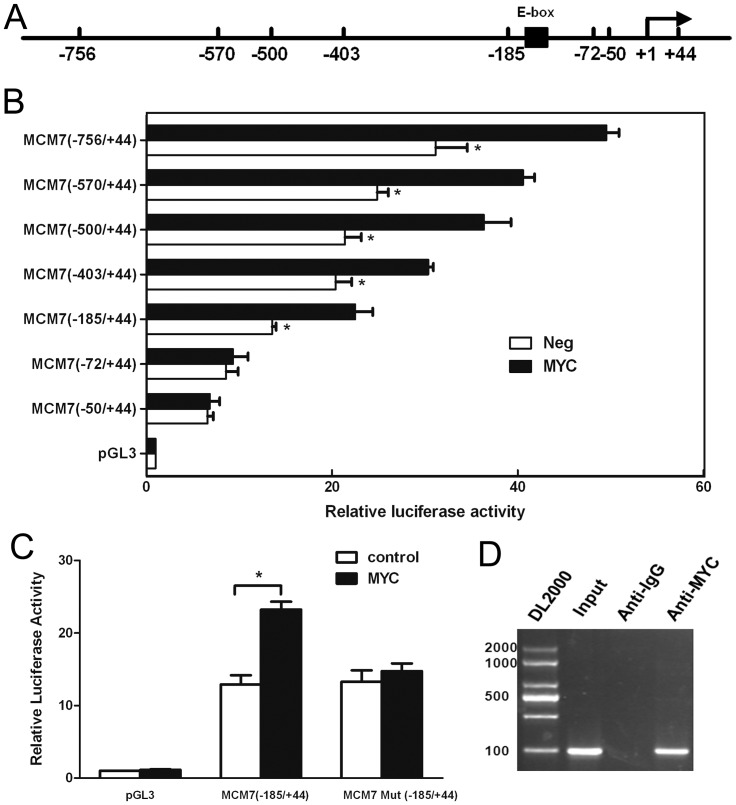
Figure 5. MYC activation of human MCM7 promoter activity was dependent on the E-box sequence.
ECC-1 cells were transfected to detect the human MCM7 promoter activity. (a) Schematic of the core promoter region of the MCM7 promoter. Location of the mutated sequence for the E box (AGTTTA) is indicated by the black box. (b) Luciferase activity of constructs of the MCM7 promoter and its truncations. ECC-1 cells transfected with luciferase reporter constructs containing different 5′flanking segments of hMCM7 alone or co-transfected with pBABE-MYC. The numbers on the left side of the panel represent the relative position of each deletion. The luciferase activities were normalized to that of ECC-1 cells transfected with the pGL3(-) vector (control) was set to 1.0. Results are means ± S.D. All assays were repeated for at least 3 independent experiments. In each experiment, the individual data points were calculated as the means of triplicates. (c) Effect of mutated E-box site on activity of the MCM7 promoter. The MCM7 (−185/+44)/luciferase reporter construct was co-transfection with its E-box site mutant and with or without the MYC expression plasmid. (d) ChIP assays were performed in ECC-1 cells using antibodies directed against MYC protein or an IgG control. The sequences of all primers for ChIP-PCR were based on the MYC-binding sites (−756 to −50 relative to the TSS) identified from TSS analysis. After immunoprecipitation, DNA was eluted and amplified by PCR using primers designed to amplify the minimal promoter region of MCM7. *, P<0.05, paired t-test.