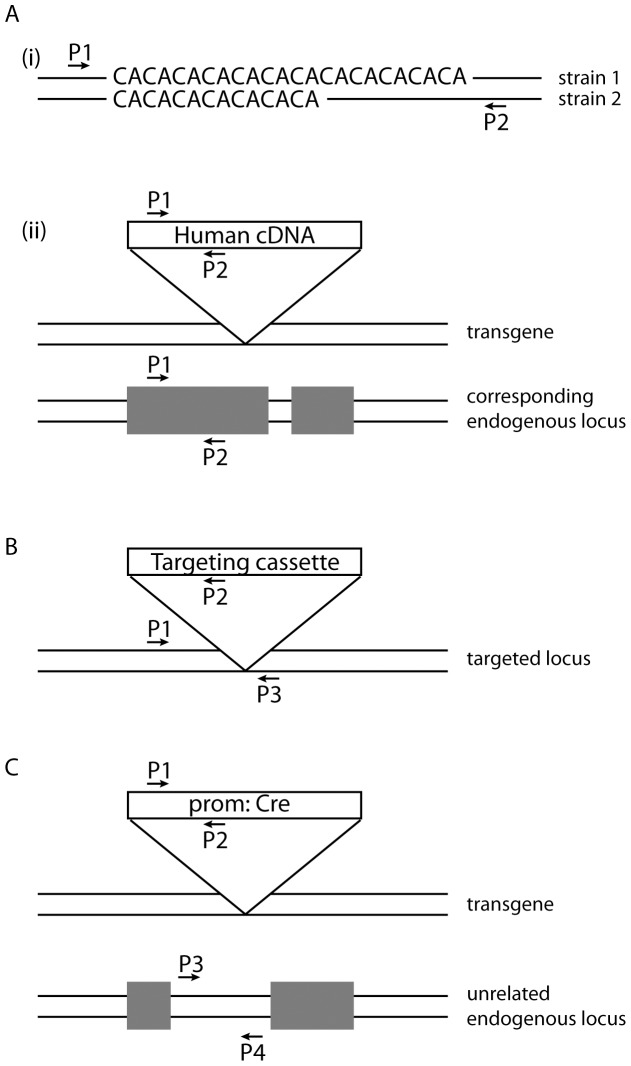
Figure 1. Plus/plus genotyping assay design.
(A) Two-primer assays are suitable when common sequences flank a divergent sequence. The divergence may for example be due to (i) strain specific polymorphisms (such as microsatellite markers), or (ii) genomic incorporation of an orthologous DNA sequence and may generate products of differing size and/or sequence. (B) Three-primer assays are appropriate when the site of incorporation of exogenous DNA is known. The P1/P2 primers will specifically amplify a product from the modified locus by virtue of the exogenous DNA specific P2. The P1/P3 primers will only amplify a product from the wild type locus because PCR conditions are set to favour the short product and inhibit the amplification of the theoretically possible but much larger product from the altered locus. (C) Four-primer assays are useful when the location of the exogenous DNA is unknown and the exogenous sequence is unrelated to the mouse genome. The P1/P2 primers are unique to the exogenous DNA and P3/P4 can correspond to any unrelated region of the mouse genome. P: primer.