Abstract
The MTERF protein family comprises members from Metazoans and plants. All the Metazoan MTERF proteins characterized to date, including the mitochondrial transcription termination factors, play a key role in mitochondrial gene expression. In this study we report the characterization of Drosophila MTERF5 (D-MTERF5), a mitochondrial protein existing only in insects, probably originated from a duplication event of the transcription termination factor DmTTF. D-MTERF5 knock-down in D.Mel-2 cells alters transcript levels with an opposite pattern to that produced by DmTTF knock-down. D-MTERF5 is able to interact with mtDNA at the same sites contacted by DmTTF, but only in the presence of the termination factor. We propose that the two proteins participate in the transcription termination process, with D-MTERF5 engaged in relieving the block exerted by DmTTF. This hypothesis is supported also by D-MTERF5 homology modeling, which suggests that this protein contains protein–protein interaction domains. Co-regulation by DREF (DNA Replication-related Element binding Factor) of D-MTERF5 and DmTTF implies that expression of the two factors needs to be co-ordinated to ensure fine modulation of Drosophila mitochondrial transcription.
Keywords: Drosophila, mtDNA transcription, Transcription termination, DmTTF, MTERF family, MTERF5
Highlights
► We characterized D-MTERF5, a Drosophila mitochondrial protein of the MTERF family. ► D-MTERF5 knock-down alters mitochondrial transcript levels. ► D-MTERF5 interacts with mtDNA at the same sites contacted by DmTTF. ► D-MTERF5 would counteract the transcription termination factor DmTTF.
1. Introduction
Mammalian mitochondrial DNA (mtDNA) is a circular molecule of about 16 kbp. Its transcription depends on two closely spaced promoters, HSP (H-strand promoter) and LSP (L-strand promoter), both located in the D-loop region. Transcription from LSP produces only one polycistronic precursor RNA, whereas transcription depending on HSP generates two partially overlapping polycistronic primary transcripts. The transcriptional unit starting from the IH1 initiation point generates 12S and 16S rRNAs and two tRNAs; the unit starting from the IH2 covers almost the entire mtDNA molecule and is processed to obtain the mRNAs for 12 polypeptides and 12 tRNAs (for review see Fernández-Silva et al., 2003; Shutt and Shadel, 2010; Peralta et al., 2011).
Nuclear-encoded proteins are responsible for transcription in mitochondria. The catalytic component of the transcription machinery consists of the phage-like single-subunit RNA polymerase POLRMT (Ringel et al., 2011). During initiation, POLRMT is assisted by two accessory factors, the HMG box protein TFAM and the rRNA methyltransferase related factor TFB2M (Litonin et al., 2010). It was recently suggested that mitochondrial transcription also requires the elongation factor TEFM, which interacts with the catalytic domain of POLRMT and enhances its processivity (Minczuk et al., 2011).
The other known factors participating in mtDNA transcription belong to a wide protein family initially named the MTERF protein family after its founder, mTERF or MTERF1 (Linder et al., 2005; Roberti et al., 2009, 2010). Vertebrates possess four paralogue genes defining four sub-families, named MTERF1, -2, -3, and -4. Sub-families MTERF1 and MTERF2 are unique to vertebrates, whereas sub-families MTERF3 and MTERF4 comprise proteins from all Metazoans. Mammalian mTERF is a sequence-specific DNA binding protein able to promote transcription termination of the IH1 transcription unit, downstream of the 16S rRNA gene (Fernandez-Silva et al., 1997). Multiple evidence suggests that mTERF arrests POLRMT progression in a bidirectional way, with a higher efficiency when POLRMT moves in the L-strand transcription direction (Asin-Cayuela et al., 2005). Martin et al. (2005) reported that mTERF is also able to promote a higher rate of transcription of rDNA by facilitating the recycling of POLRMT between the termination site and the H1 initiation site (Martin et al., 2005). The other mammalian MTERF proteins have also been characterized. MTERF2, which likely originated together with mTERF from a duplication event that occurred in the vertebrate ancestor lineage, plays a role in transcription, as well as MTERF3. Both interact with the mtDNA promoter region with MTERF3 acting as a repressor and MTERF2 being a positive modulator (Park et al., 2007; Wenz et al., 2009). Moreover, it has also been reported that alterations in the level of MTERF1, MTERF2 and MTERF3 cause impairment of the mtDNA replication process (Hyvärinen et al., 2007, 2011). Mammalian MTERF4 has been very recently characterized. This protein participates in ribosome assembly rather than transcription (Cámara et al., 2011).
Invertebrate mitochondrial genomes share the same gene content of mammals. However, they show profound variations in gene organization, probably reflecting differences in the mechanisms of transcription. In particular, the mtDNA from Drosophila melanogaster is about 19.5 kbp long, with an AT-rich non-coding region accounting for the larger size of the genome (Lewis et al., 1995). In contrast to mammals, in Drosophila genes are almost equally distributed between the two strands of mtDNA, and form four clusters that are located alternatively on the two strands.
Concerning transcription factors, Drosophila possesses evident orthologues of mammalian TFAM and TFB2M (Adán et al., 2008; Takata et al., 2001). The transcription termination factor DmTTF has also been characterized (Roberti et al., 2003). The protein, belonging to the MTERF family, recognizes two homologous sequences on mtDNA that are placed at the boundary of gene clusters transcribed in opposite directions, namely the boundaries between the tRNAGlu and tRNAPhe genes and between the tRNASer(UCN) and ND1 genes. The role of DmTTF as a bidirectional transcription termination factor has been extensively addressed in vitro and in vivo (Roberti et al., 2005, 2006, 2009). In particular, DmTTF knock-down and over-expression in D.Mel-2 cells cause substantial alteration of the steady-state level of mitochondrial sense and antisense transcripts, consistently with the ability of the protein to arrest the progression of POLRMT.
Besides DmTTF, D. melanogaster possesses three more MTERF proteins (Linder et al., 2005; Roberti et al., 2010). They are the orthologues of mammalian MTERF3 and MTERF4 and a polypeptide reported in FlyBase Database as CG7175. In this paper, we report for the first time the functional characterization of CG7175 (here renamed as D-MTERF5), by a combination of in vivo and in vitro approaches. The obtained data indicate that this polypeptide and DmTTF interact with the same sites on mtDNA and play antithetical roles in mitochondrial transcription termination. This would allow the fine tuning of transcription in Drosophila mitochondria.
2. Materials and methods
2.1. Cloning of D-MTERF5 cDNA
D-MTERF5 cDNA was obtained by both PCR amplification of a D. melanogaster cDNA library from 2–14 hour embryos and RT-PCR on total RNA from S2 cells. Primers were CG7175-UTR-For (TCTGCGGGAAATGTTATAATT), nt 231–251 of the mRNA sequence (GenBank ID: NM_142436) CG7175-For (ATGCTGCGCAACGGCAGAAT), nt 252–272, and CG7175-Rev (CTACAAGCGCTCCATAATCATTTT), nt 1934–1911. The amplification product (1683 bp) was cloned into the vector pCR®2.1-TOPO® (Invitrogen) and sequenced.
2.2. Drosophila cell culture and conditions for RNAi
Drosophila embryonic D.Mel-2 cells were maintained at 28 °C in HyQ SFM (HyClone) supplemented with 16 mM l-glutamine, 50 units/ml penicillin and 50 μg/ml streptomycin. For RNAi treatment, cells were diluted to a concentration of 1.0 × 106 cells/ml in 10 ml of complete HyQ SFM in 75-cm2 flasks. dsRNA (15 μg per 106 cells) was added directly to the medium. Flasks were swirled by hand and cells were incubated at 28 °C for 1 h. Then, 10 ml of medium was added to obtain a cell density of 0.5 × 106 cells/ml, an additional incubation at 28 °C for 72 h followed. D-MTERF5 and Opa1 dsRNAs were produced using the MEGAscript T7 transcription kit (Ambion). Templates were PCR-derived fragments carrying the T7 promoter sequence at both ends. The gene-specific sequences of the primers were as follows: D-MTERF5 (GenBank ID: NM_142436) forward-primer nt 269–284, reverse-primer nt 1054–1040; Opa1 (GenBank ID: NM_079504) forward-primer nt 303–317, reverse-primer nt 1017–1004. DREF RNAi was performed as reported by Fernández-Moreno et al. (2009).
2.3. Subcellular localization of D-MTERF5 by confocal microscopy
D.Mel-2 cells were diluted to a final concentration of 2.0 × 106 cells/ml in 5 ml of antibiotic-free HyQ SFM in 75-cm2 flasks 16 h prior to transfection. Ten micrograms of plasmid pMt/Hy containing the entire coding sequence of D-MTERF5 fused with the FLAG-tag sequence was added in the presence of 50 μg of Cellfectin (Invitrogen) in fresh antibiotic-free medium. Cells were incubated at 28 °C for 24 h and then induced with 500 μM CuSO4. Twenty hours after induction, cells were settled down on poly l-lysine coated glass coverslips and mitochondria were stained with 250 nM MitoTracker Red CMXRos (Invitrogen-Molecular Probes) at 20 °C for 1 h in Drosophila SFM medium. Then cells were washed with phosphate buffered saline (PBS), fixed with 3.7% formaldehyde/PBS for 20 min and permeabilized with 0.1% Triton X-100 for 10 min. After blocking with 0.1% gelatin in PBS for 45 min, coverslips were incubated with mouse monoclonal anti-FLAG M2 antibody (Sigma, 1:500 dilution in 0.1% gelatin in PBS) at room temperature for 2 h, washed three times for 5 min each with 0.1% gelatin in PBS and incubated with an Alexa Fluor 488 conjugated anti-mouse antibody (Invitrogen-Molecular Probes, 1:1000 dilution in 0.1% gelatin in PBS) at room temperature for 1 h. Finally, coverslips were washed three times with PBS, rapidly rinsed in distilled water, and then mounted on glass slides with glycerol/PBS. Confocal images were captured by means of a Leica TCS SP5 confocal microscope (Leica Microsystems, Mannheim, Germany) using a 63 × oil immersion objective (N.A. = 1.32) and a 100 mW Argon laser (488 nm and 543 nm lines).
2.4. RT-PCR and real-time RT-PCR assays
Total cellular RNA was extracted from treated and untreated D.Mel-2 cells by the RNeasy Midi Kit (Qiagen). Semiquantitative RT-PCR assay was performed using the Enhanced Avian HS RT-PCR Kit (Sigma). Reactions contained 300–500 ng of Drosophila total RNA as template, and 50 pmol of sequence specific primers in a final volume of 50 μl.
For real-time RT-PCR assay, RNA was reverse-transcribed using the Enhanced AMV Reverse Transcriptase (Sigma). Each reaction was carried out in a final volume of 25 μl using 250 ng of total RNA and 25 pmol of mtDNA gene-specific primer. Real-time PCR was performed using the SYBR® Green PCR MasterMix (Applied Biosystems). Primers were designed using the Primer Express 2.0 software (Applied Biosystems) and sequence positions on D. melanogaster mtDNA (GenBank ID: NC_001709) were as follows: ND2-for nt 558–573, ND2-rev nt 651–632; COI-for nt 1838–1862, COI-rev nt 1932–1904; COII-for nt 3569–3593, COII-rev nt 3652–3631; COIII-for nt 5304–5328, COIII-rev nt 5379–5355; ND5-for nt 7040–7061, ND5-rev nt 7114–7095; ND4/4L-for nt 8561–8584, ND4/4L-rev nt 8722–8696; cyt b-for nt 10697–10719, cyt b-rev nt 10766–10748; ND1-for nt 11984–12002, ND1-rev nt 12056–12034;lrRNA-for nt 12827–12845, lrRNA-rev nt 12906–12887. RP49 rRNA (GenBank ID: NM_170461) and 28S rRNA (GenBank ID: M21017) were used as an endogenous control; primer sequence positions were: RP-for nt 189–208, RP-rev nt 319–301 and 28S-for nt 1407–1429, 28S-rev nt 1480–1462, respectively. Real-time RT-PCR reactions and analysis were conducted as described by Roberti et al. (2006).
2.5. Protein extract preparation and Western blotting analysis
Preparation of mitochondrial protein extracts from D.Mel-2 cells, as well as Western blotting detection of DmTTF, was carried out as described by Roberti et al. (2006). For Western blotting analysis of D-MTERF5, mitochondrial proteins were electrophoresed on 12% SDS-polyacrylamide gel and electro-transferred to PVDF (Hybond-P, GE Healthcare). Membranes were pre-incubated in 1 × PBS, 3% BSA for 1 h, and then incubated in the same solution for 16 h at 4 °C in the presence of polyclonal antibodies raised in rabbit against recombinant D-MTERF5 (Eurogentec). Membranes were then washed twice in 1 × PBS, 3% BSA and twice in 1 × PBS and incubated for 1 h with horseradish peroxidase-conjugated anti-rabbit IgG (Santa Cruz Biotechnology) and washed with 1 × PBS. Protein bands were visualized using the ECL Plus Western blotting reagents (GE Healthcare).
2.6. Production of recombinant proteins and binding assays
The mature form of DmTTF was expressed in bacteria using the pCR® T7 TOPO TA Expression System (Invitrogen) and purified by Heparin-Sepharose chromatography as reported in Roberti et al. (2003).
For expression in bacteria, the mature version of D-MTERF5 (aa 30–560) was cloned in the pCR® T7/CT TOPO vector (Invitrogen) with a 6xHis-tag at its C-terminus. Exponentially growing Escherichia coli BL21(DE3)pLysS cells transformed with the recombinant plasmid were induced with IPTG for 2 h at 18 °C. Cells were resuspended in 50 mM potassium phosphate, pH 7.8, 400 mM NaCl, 100 mM KCl, 10% glycerol, 0.5% Triton X-100 and sonicated. The bacterial lysate was loaded onto HisTrapTM HP affinity column (GE Healthcare) equilibrated with buffer A (25 mM potassium phosphate, pH 7.4, 500 mM NaCl, 10% glycerol) containing 30 mM imidazole. After a wash with buffer A containing 150 mM imidazole, D-MTERF5 was eluted with buffer A containing 500 mM imidazole. EMSA and footprinting assays were performed as previously described (Roberti et al., 2003), with the only exception that DNA probes were fluorescently labeled using modified PCR primers carrying at their 5′-end the cyanine5 group.
3. Results and discussion
3.1. D-MTERF5 is a Drosophila mitochondrial protein of the MTERF family
We focused our attention on the MTERF family protein of D. melanogaster annotated as CG7175 in the FlyBase Database. The CG7175 gene is 2160 bp long, comprises four exons and three introns and generates a transcript of 1998 nt codifying for a 560 residue polypeptide. This protein appears more closely related to the transcription termination factor DmTTF (14% of identity and 42% of similarity), than to the other Drosophila MTERF proteins (13% identity and 24% similarity with respect to MTERF3; 7% identity and 20% similarity with respect to MTERF4). Similarly to DmTTF, CG7175 does not belong to any of the four MTERF sub-families (Linder et al., 2005). Since orthologues of this protein are found only in insects, it could be argued that in the insect lineage an early gene duplication event occurred that in D. melanogaster generated the transcription termination factor DmTTF and the CG7175 polypeptide. From now on the CG7175 polypeptide will be named D-MTERF5.
MitoProt II software predicted that D-MTERF5 is imported into mitochondria with high probability (99.4 score) and possesses a signal sequence of 29 residues. Therefore the mature form of the protein (531 amino acid long) should have a calculated molecular weight of 61.8 kDa and an isoelectric point of 8.78.
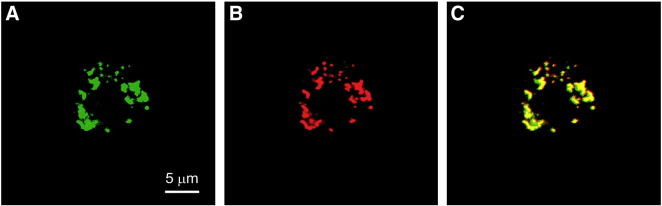
To verify the mitochondrial localization of the protein, we over-expressed the C-terminal FLAG-tagged D-MTERF5 in D.Mel-2 cells and analyzed its overlap with the Mito Tracker dye by immunocytochemistry. The confocal microscopy image shown in Fig. 1 clearly demonstrates that FLAG-tagged D-MTERF5 co-localizes with the mitochondrial network.
Fig. 1.
D-MTERF5 is a mitochondrial protein. Confocal microscopy on D.Mel-2 cells over-expressing FLAG-tagged D-MTERF5 and stained with anti-FLAG mouse antibody plus Alexa Fluor 488 conjugated anti-mouse antibody (green, panel A) and with MitoTraker (red, panel B). Co-localization of the green and red signals is showed in the merged image (yellow, panel C).
3.2. D-MTERF5 influences mitochondrial transcription
To obtain information on the role of D-MTERF5, a protein knock-down phenotype was produced by treating D.Mel-2 cells with a dsRNA encompassing 787 nt of the D-MTERF5 coding sequence. Depletion of D-MTERF5 mRNA was monitored by semiquantitative RT-PCR assay using primers delimiting the entire coding sequence (about 1680 bp). The level of D-MTERF5 mRNA in treated cells was almost undetectable up to 40 amplification cycles (Fig. 2A); the decrease was specific since no effect was obtained in mock control cells treated with dsRNA containing the sequence of the Opa1 gene (not shown).
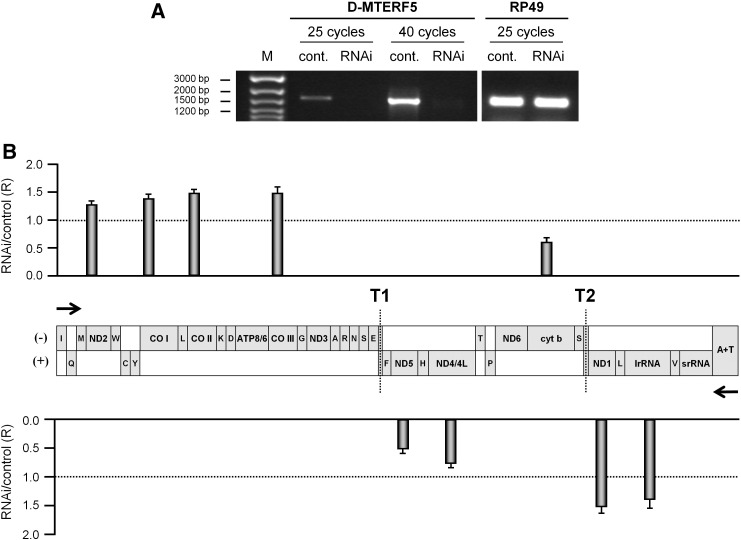
Fig. 2.
D-MTERF5 depletion affects mitochondrial transcription. (A) D-MTERF5-targeted RNAi in D.Mel-2 cells was monitored by RT-PCR. Total RNA (800 ng) extracted from cells untreated (cont.), treated with D-MTERF5 dsRNA (RNAi) was used as template in RT-PCR reactions. Samples were run on a 1.5% agarose gel and stained with ethidium bromide. Nuclear-encoded RP49 mRNA was used as endogenous control. (B) Relative quantification of mitochondrial transcripts by real-time RT-PCR. The mtDNA molecule is represented as a linear map. The directions of transcription of the (−) and (+) strands are indicated by black arrows; the two DmTTF binding sites are marked by vertical dotted lines and are indicated as T1 (the site at the boundary between the tRNAGlu and tRNAPhe genes) and T2 (the site at the boundary between the tRNASer(UCN) and ND1 genes). Bars reported above the (−) strand and below the (+) strand, in correspondence to the position of the analyzed genes, indicate the relative content (R) of transcript, normalized to RP49 mRNA (endogenous control), in D-MTERF5 depleted (RNAi) with respect to control cells fixed as 1-value. The relative quantification was performed according to the equation R = (ET)ΔCt,T / (EC)ΔCt,C (Pfaffl, 2001). For each amplicon, the mean ratio value was obtained from at least four real-time RT-PCR assays, using RNA obtained from independent RNAi experiments; standard deviations are indicated on the top of the bars. Statistical analysis, performed using paired two-tailed Student's t-test showed that the differences between RNAi and control are all significant (P < 0.05).
Since D-MTERF5 belongs to the MTERF family, whose members include factors controlling mtDNA transcription and replication, we decided to test the effect of a D-MTERF5 knock-down on mtDNA content and mitochondrial transcript levels. MtDNA copy number, determined as a mtDNA/nDNA ratio by means of real-time PCR, was found not to vary significantly between control and RNAi cells (data not shown).
To investigate the effect of D-MTERF5 knock-down on mitochondrial transcription, we measured the steady-state level of nine RNAs distributed on both mtDNA strands. We found that the level of all analyzed transcripts varied significantly in depleted cells (Fig. 2B). In particular, the (−) strand transcripts ND2, COI, COII and COIII increased to about 30–50%, whereas cyt b mRNA decreased to about 40%. The (+) strand transcripts lrRNA and ND1 showed an increased level (40% and 50%, respectively); ND4/4L and ND6 mRNAs showed a 30% and 50% decrease, respectively.
To rule out the possibility that D-MTERF5 knock-down could have altered the levels of other proteins involved in mtDNA transcription and in turn, the abundance of mitochondrial transcripts, we measured the mRNA level of POLRMT, the initiation factors TFAM and TFB2M, the repressor MTERF3 and the termination factor DmTTF. In all cases we found no variations in RNAi cells with respect to control cells (data not shown); hence, we conclude that the observed changes in mitochondrial RNA levels are likely due to a direct consequence of the D-MTERF5 knock-down. Altogether these data strongly support the involvement of D-MTERF5 in Drosophila mitochondrial transcription.
Importantly, a clear correlation can be observed between the trend in the variation of transcript levels in DMTERF5 depleted cells and the location of DmTTF binding sites (see Fig. 2B and Roberti et al, 2003). Both the levels of the ND2, COI, COII and COIII transcripts in the (−) strand and the IrRNA and ND1 transcripts in the (+) strand, which, according to transcription direction, map upstream of the T1 and T2 DmTTF binding sites, respectively, increased upon knock-down of D-MTERF5. Conversely, levels of the cyt b mRNA in the (−) strand and the ND4/4L and ND5 mRNAs in the (+) strand, which map downstream of T1 and T2, respectively, decreased when D-MTERF5 is knocked down. Since, as we previously reported (Roberti et al, 2006) in DmTTF depleted cells, on both mtDNA strands, transcripts mapping upstream of the T1 and T2 binding sites were decreased, whereas transcripts mapping downstream of the protein binding sites were increased, the effect of D-MTERF5 RNAi on transcript levels is exactly the opposite to that produced by DmTTF knock-down. Therefore, the role of D-MTERF5 in transcription appears to be somehow antithetical to that of the transcription termination factor.
In the light of the possible involvement of D-MTERF5 in the DmTTF mediated termination process, we measured the in vivo levels of both proteins in D.Mel-2 cells by Western blot (Supplementary Fig. 1) and we found that the contents of D-MTERF5 and DmTTF were 0.8 ± 0.17 pmol and 2.3 ± 0.25 pmol per mg of total mitochondrial proteins, respectively. Therefore the amount of D-MTERF5 appears to be in the same order of magnitude of that of the transcription termination factor.
3.3. D-MTERF5 interacts with DmTTF binding sites on mtDNA
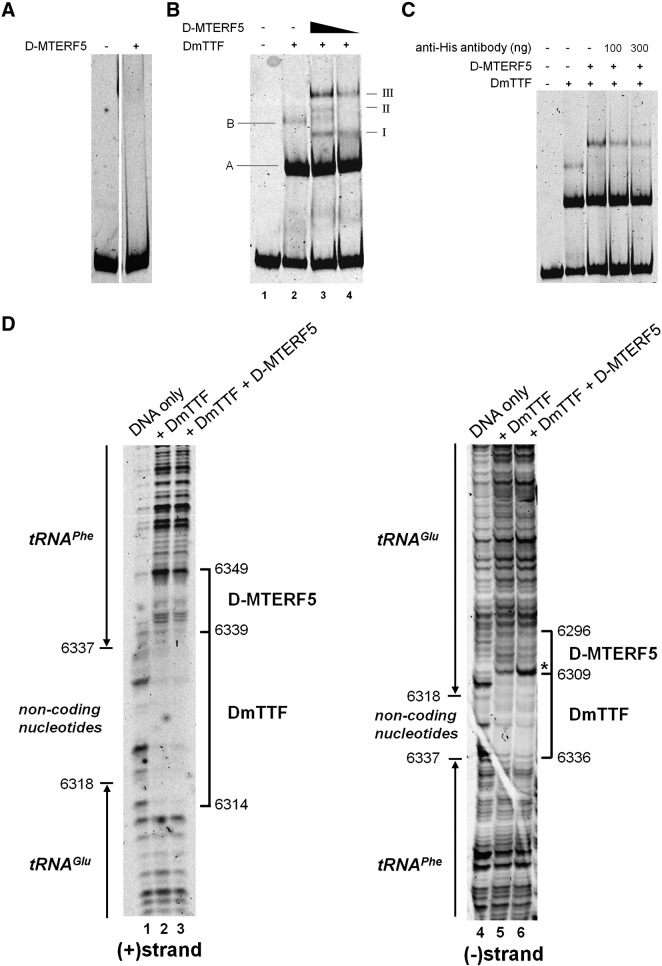
The above data prompted us to investigate the possibility that D-MTERF5 could interact with mtDNA at the same sites bound by DmTTF. We expressed a His-tagged mature form of D-MTERF5 in bacteria, and purified it at almost homogeneity by means of metal-affinity chromatography (Supplementary Fig. 2A). We first tested the capacity of recombinant D-MTERF5 to interact in an EMSA assay with a 139 bp fragment, which spans the tRNAGlu and tRNAPhe genes almost completely, and contains the DmTTF T1 binding site. As reported in Fig. 3A, no evident retarded complexes were obtained in the presence of a large excess of protein, thus arguing against the capacity of D-MTERF5 to bind this mtDNA region per se. On the other hand, the possible involvement of DmTTF and D-MTERF5 in the same process suggested that a physical interaction could occur between the two proteins. On this basis, and considering the nearly stoichiometric level of the two proteins observed in vivo, we investigated the binding of D-MTERF5 to the EMSA probe in the presence of recombinant purified DmTTF (see Supplementary Fig. 2B, for DmTTF purification). The results of the mobility shift experiment are shown in Fig. 3B. In the presence of DmTTF only (lane 2), we observed the typical DmTTF–DNA complexes with the faster migrating band (A) representing the most abundant complex formed by one DmTTF molecule, and the slower migrating band (B) consisting in the less abundant complex likely formed by two DmTTF molecules. When D-MTERF5 was added to the binding reaction, we observed additional retarded bands migrating more slowly than the DmTTF/DNA complex A; the abundance of these bands was dependent on the amount of D-MTERF5 (lanes 3 and 4, complexes I–III). Electrophoretic mobility suggests that complex I is generated by D-MTERF5 alone and complexes II and III contain both proteins, probably in various stoichiometries. Differently from complexes I and II, which resulted as poorly reproducible (see Fig. 3C) probably due to their low stability, complex III was the most abundant and always reproducible. The presence of D-MTERF5 in complex III was confirmed by using anti-His antibodies (Fig. 3C). Increasing amounts of antibodies added to the reaction mixture interfered with the binding of His-tagged-D-MTERF5 to DmTTF–DNA complex and caused the progressive decrease of complex III formation.
Fig. 3.
Analysis of the DNA-binding properties of D-MTERF5. EMSA and footprinting assays were employed to test the interaction of D-MTERF5 with mtDNA at the tRNAGlu/tRNAPhe boundary region containing the T1 DmTTF binding site. A DNA fragment spanning position nt 6261–6399 of D. melanogaster mtDNA (GenBank ID: NC_001709) labeled with cyanine5 at the 5′-ends was used as a probe in the binding reactions. (A) EMSA was conducted by incubating the DNA probe (25 fmol) with 50 ng (about 0.8 pmol) of recombinant purified D-MTERF5. (B) EMSA was conducted by incubating the DNA probe (25 fmol) with 25 or 12.5 ng (about 0.4 and 0.2 pmol, respectively) of recombinant purified D-MTERF5 in the presence of 50 ng (about 1.2 pmol) of recombinant purified DmTTF. Complexes formed by DmTTF are indicated by capital letters, and complexes obtained following the addition of D-MTERF5 are indicated by Roman numerals. (C) The indicated amounts of anti-6xHis antibodies were added to the EMSA reaction mixture containing 25 fmol of DNA and 50 ng of both D-MTERF5 and DmTTF (about 0.8 and 1.2 pmol, respectively). (D) DNase I protection analysis was conducted on protein–DNA complexes obtained by adding to 100 fmol of DNA fragment labeled at the 5′-end of either (+) or (−) strand, 500 ng (about 8.2 pmol) of purified DmTTF or 500 ng of DmTTF (about 8.2 pmol) plus 250 ng of D-MTERF5 (about 4.0 pmol). Protected regions are indicated by brackets; the asterisk indicates the DNase I-hypersensitive site.
On the basis of the EMSA results we investigated the capacity of D-MTERF5 to directly contact mtDNA in the presence of DmTTF by means of DNaseI footprinting experiments. When the EMSA probe labeled in the (+) strand was incubated with both DmTTF and D-MTERF5 (Fig. 3D, lane 3), the typical protection by DmTTF (6314–6339) was extended by about 10 nucleotides in the direction of the tRNAPhe gene. When the DNA fragment was labeled in the (−) strand, addition of D-MTERF5 (Fig. 3D, lane 6) caused a weak protection extending by about 15 nucleotides in the direction of the tRNAGlu gene. A clear hypersensitivity site at the boundary of the DmTTF footprint was also observed. The obtained data indicate that D-MTERF5 modifies the DNaseI protection pattern produced by DmTTF.
3.4. Homology modeling of D-MTERF5
Recently the crystal structure of mTERF and its binding mode to mtDNA have been clarified (Jiménez-Menéndez et al., 2010; Yakubovskaya et al., 2010). The protein shows a modular architecture due to the repetition of eight-nine 35-residue mterf motifs, consisting of three short alpha-helices forming a left handed triangular superhelix folded around a hydrophobic core. The motifs form a solenoid-like structure, which twists to the right acquiring a convex surface and a positively charged concave surface that accommodates the DNA duplex. Sequence specific interactions with DNA cause protein and DNA conformational changes that are necessary for promoting transcription termination. The crystal structure of human MTERF3 has been also solved (Spåhr et al., 2010). It is very similar to that of mTERF and consists in the repetition of seven mterf motifs; however, MTERF3 lacks the residues that in mTERF are involved in sequence-specific DNA binding.
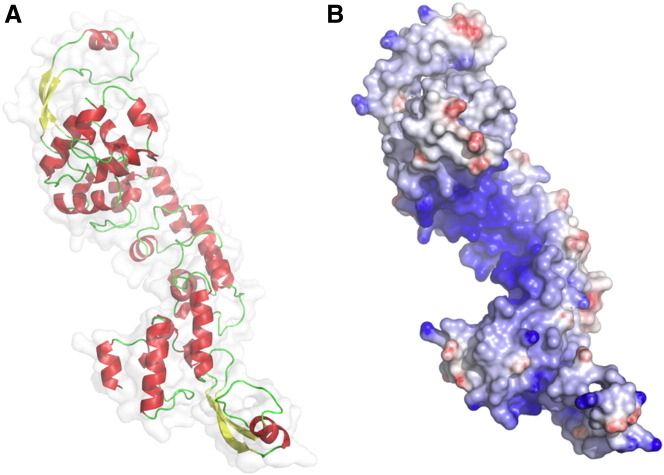
The primary structure of D-MTERF5 also contains several repetitions of the mterf motif. Therefore, it is highly probable that the protein adopts a three-dimensional shape similar to that determined for human mTERF and MTERF3. Homology modeling of D-MTERF5 performed using the Swiss-Model server confirmed this hypothesis, showing that the predicted structure of the protein is arranged as the mTERF-like series of left handed super-helices forming a right twisted solenoid structure (Fig. 4A). D-MTERF5 would also possess the positively charged concave surface able to accommodate the negatively charged double helix, being therefore, in principle, able to bind DNA (Fig. 4B). Interestingly, the predicted structure shows the presence, in both the N- and C-terminal halves of the protein, of a beta–alpha–beta domain protruding from the convex surface, which interrupts the series of super-helices. The position of the predicted mterf motifs and beta-alpha-beta domains is depicted on the D-MTERF5 sequence reported in Supplementary Fig. 3.
Fig. 4.
Homology modeling of D-MTERF5. (A) Overall structure of D-MTERF5 (residues 62–443 of the mature protein) obtained by comparison with human mTERF, by using the Swiss-Model Automated Protein Modeling Server, in which predicted alpha-helices are in red and beta-sheets are in yellow. (B) Molecular surface of D-MTERF5 colored according to the electrostatic surface potential (blue + 12 kT, red − 12 kT).
It is known that beta-sheet domains can be used by proteins (as homo- or hetero-dimerization domains) to establish contacts with interacting partners (Remaut and Waksman, 2006); therefore, it can be supposed that D-MTERF5, at the termination sites, could use the beta–alpha–beta domains to establish interactions with other proteins, such as POLRMT or the termination factor. Interestingly, structure modeling suggests that DmTTF also contains two short beta-sheet domains (not shown).
3.5. D-MTERF5 expression is controlled by DREF
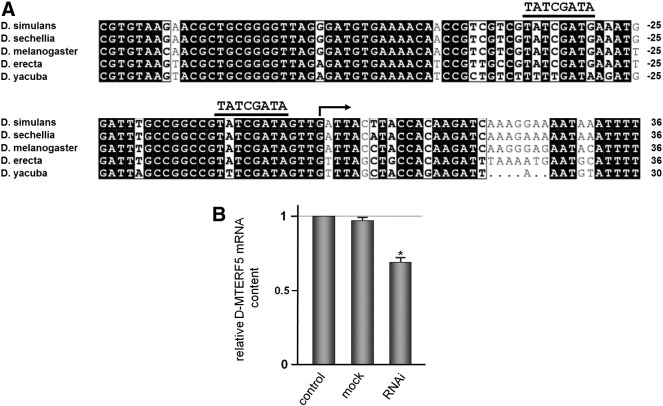
We analyzed the control of D-MTERF5 gene expression by DREF (DNA Replication-related Element binding Factor). DREF is a nuclear factor that activates genes involved in nuclear DNA replication and cell cycle control (Matsukage et al., 2007), and is also able to activate genes involved in mtDNA replication and maintenance (Lefai et al., 2000; Ruiz De Mena et al., 2000; Takata et al., 2001), as well as the DmTTF gene (Fernández-Moreno et al., 2009). We found DRE sequences in the D-MTERF5 promoter at positions-11 and -37; they showed 8/8 and 7/8 matches with respect to the DRE consensus TATCGATA and were highly conserved in other Drosophila species (Fig. 5A). We investigated the effect of DREF depletion on the level of D-MTERF5 transcript. DREF knock-down (more than a 90% decrease of the polypeptide level) was obtained by RNAi treatment in Drosophila S2 cultured cells, as already reported by Fernández-Moreno et al. (2009). The results from three independent RNAi experiments indicated that in DREF-depleted cells the steady-state level of D-MTERF5 mRNA, measured by real-time RT-PCR, is diminished to about 70% with respect to non-depleted cells (Fig. 5B), thus demonstrating a positive control by DREF on D-MTERF5 expression. Co-regulation by DREF of DmTTF and D-MTERF5 suggests that expression of the two factors needs to be co-ordinated to ensure modulation of Drosophila mitochondrial transcription.
Fig. 5.
Regulation of D-MTERF5 expression by DREF. (A) Multialignment of D-MTERF5 proximal promoters from different Drosophila species showing the location and conservation of DRE sequences. Promoter nucleotide sequences were obtained from FlyBase Database. ClustalW alignment was performed at NPS@ web server of the PBIL. The arrow indicates the transcription start point according to the D. melanogaster + 1 site as in FlyBase. DRE sequences within the promoters are indicated by solid bars; the DRE consensus sequence is also shown above bars. (B) Effect of DREF knock-down on D-MTERF5 mRNA levels. DREF protein was knocked down in S2 cells by means of RNAi as reported by Fernández-Moreno et al. (2009). Total RNA was extracted from Drosophila S2 cells, either untreated (control) or treated with LacZ (mock) or DREF dsRNA; relative quantification of D-MTEF5 mRNAs was carried out by real-time RT-PCR. Bars indicate the relative content of transcript, normalized to 28S rRNA (endogenous control), with that in control cells fixed as 1-value. Relative quantification and statistical analysis were performed as described in the Fig. 2 legend (* indicates P < 0.05).
3.6. Role of D-MTERF5 in transcription
The results of DNA-binding assays and homology modeling allow us to propose at least two possible models to explain the effect of D-MTERF5 on mitochondrial transcription. According to the first one, the protein would bind DNA with no sequence specificity and very low affinity; however, when bound to DmTTF, D-MTERF5 would acquire the capacity to specifically contact mtDNA at the termination sites. Alternatively, D-MTERF5 could function not by directly binding DNA, but by contacting DmTTF only and inducing in its structure conformational changes that could modify the interaction of DmTTF with DNA. In either case, the symmetric expansion of the DmTTF footprint produced by D-MTERF5 suggests that it is able to bind the DmTTF–DNA complex on both ends, being this is in line with the bidirectional termination activity of DNA-bound DmTTF (Roberti et al., 2005). Whatever the nature of the interactions between D-MTERF5 and the DmTTF/DNA complex, the protein would negatively affect the capacity of the transcription termination factor to arrest POLRMT, thus explaining the enhanced transcription termination activity observed in D-MTERF5 depleted cells. Therefore, by acting in combination with DmTTF, D-MTERF5 might be a key modulator of Drosophila mitochondrial transcription.
Although the mtDNA transcription mechanism in flies still remains uncertain, the most likely model assumes that transcription of each strand depends on one promoter located in the A + T-rich main non-coding region (Roberti et al., 2009). According to this model, the two DmTTF–DNA complexes would mainly serve to coordinate the passage of transcription machineries moving on opposite strands. In this scenario, the role of D-MTERF5 would be to relieve the block to POLRMT imposed by DmTTF, thus permitting the enzyme to transcribe the genes mapping downstream of the transcription termination sites. Therefore, the appearance of MTERF5 function in insects could be related to the peculiar gene organization of their mtDNA and would allow the fine tuning of transcription in insect mitochondria.
Acknowledgments
This work was supported by grants from: Università di Bari, Progetto di Ricerca di Ateneo, M.I.U.R. COFIN-PRIN 2008 and Telethon — Italy (grant GGP06233). Authors thank T. De Filippis (Dept. of Biosciences, Biotech. and Pharmacol. Sciences, Univ. of Bari) for her precious technical assistance and F. Fracasso (Dept. of Biosciences, Biotech. and Pharmacol. Sciences, Univ. of Bari) for his help in preparing figures and manuscript.
Appendix A. Supplementary data
Supplementary materials.
References
- Adán C., Matsushima Y., Hernández-Sierra R., Marco-Ferreres R., Fernández-Moreno M.A., González-Vioque E., Calleja M., Aragón J.J., Kaguni L.S., Garesse R. Mitochondrial transcription factor B2 is essential for metabolic function in Drosophila melanogaster development. J. Biol. Chem. 2008;283:12333–12342. doi: 10.1074/jbc.M801342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asin-Cayuela J., Schwend T., Farge G., Gustafsson C.M. The human mitochondrial transcription termination factor (mTERF) is fully active in vitro in the non-phosphorylated form. J. Biol. Chem. 2005;280:25499–25505. doi: 10.1074/jbc.M501145200. [DOI] [PubMed] [Google Scholar]
- Cámara Y., Asin-Cayuela J., Park C.B., Metodiev M.D., Shi Y., Ruzzenente B., Kukat C., Habermann B., Wibom R., Hultenby K., Franz T., Erdjument-Bromage H., Tempst P., Hallberg B.M., Gustafsson C.M., Larsson N.-G. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527–539. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Fernández-Moreno M.A., Bruni F., Adán C., Hernandez Sierra R., Loguercio Polosa P., Cantatore P., Garesse R., Roberti M. The Drosophila nuclear factor DREF positively regulates the expression of the mitochondrial transcription termination factor DmTTF. Biochem. J. 2009;418:453–462. doi: 10.1042/BJ20081174. [DOI] [PubMed] [Google Scholar]
- Fernandez-Silva P., Martinez-Azorin F., Micol V., Attardi G. The human mitochondrial transcription termination factor (mTERF) is a multizipper protein but binds to DNA as a monomer, with evidence pointing to intramolecular leucine zipper interactions. EMBO J. 1997;16:1066–1079. doi: 10.1093/emboj/16.5.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Silva P., Enriquez J.A., Montoya J. Replication and transcription of mammalian mitochondrial DNA. Exp. Physiol. 2003;88:41–56. doi: 10.1113/eph8802514. [DOI] [PubMed] [Google Scholar]
- Hyvärinen A.K., Pohjoismäki J.L., Reyes A., Wanrooij S., Yasukawa T., Karhunen P.J., Spelbrink J.N., Holt I.J., Jacobs H.T. The mitochondrial transcription termination factor mTERF modulates replication pausing in human mitochondrial DNA. Nucleic Acids Res. 2007;35:6458–6474. doi: 10.1093/nar/gkm676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvärinen A.K., Pohjoismäki J.L., Holt I.J., Jacobs H.T. Overexpression of MTERFD1 or MTERFD3 impairs the completion of mitochondrial DNA replication. Mol. Biol. Rep. 2011;38:1321–1328. doi: 10.1007/s11033-010-0233-9. [DOI] [PubMed] [Google Scholar]
- Jiménez-Menéndez N., Fernández-Millán P., Rubio-Cosials A., Arnan C., Montoya J., Jacobs H.T., Bernadó P., Coll M., Usón I., Solà M. Human mitochondrial mTERF wraps around DNA through a left-handed superhelical tandem repeat. Nat. Struct. Mol. Biol. 2010;17:891–893. doi: 10.1038/nsmb.1859. [DOI] [PubMed] [Google Scholar]
- Lefai E., Fernández-Moreno M.A., Alahari A., Kaguni L.S., Garesse R. Differential regulation of the catalytic and accessory subunit genes of Drosophila mitochondrial DNA polymerase. J. Biol. Chem. 2000;275:33123–33133. doi: 10.1074/jbc.M003024200. [DOI] [PubMed] [Google Scholar]
- Lewis D.L., Farr C.L., Kaguni L.S. Drosophila melanogaster mitochondrial DNA: completion of the nucleotide sequence and evolutionary comparisons. Insect Mol. Biol. 1995;4:263–278. doi: 10.1111/j.1365-2583.1995.tb00032.x. [DOI] [PubMed] [Google Scholar]
- Linder T., Park C.B., Asin-Cayuela J., Pellegrini M., Larsson N.-G., Falkenberg M., Samuelsson T., Gustafsson C.M. A family of putative transcription termination factors shared amongst metazoans and plants. Curr. Genet. 2005;48:265–269. doi: 10.1007/s00294-005-0022-5. [DOI] [PubMed] [Google Scholar]
- Litonin D., Sologub M., Shi Y., Savkina M., Anikin M., Falkenberg M., Gustafsson C.M., Temiakov D. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J. Biol. Chem. 2010;285:18129–18133. doi: 10.1074/jbc.C110.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Cho J., Cesare A.J., Griffith J.D., Attardi G. Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell. 2005;123:1227–1240. doi: 10.1016/j.cell.2005.09.040. [DOI] [PubMed] [Google Scholar]
- Matsukage A., Hirose F., Yamaguchi M., Yoo M.-A. The DRE/DREF transcription regulatory system: a master key for cell proliferation. Biochim. Biophys. Acta. 2007;1779:81–89. doi: 10.1016/j.bbagrm.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Minczuk M., He J., Duch A.M., Ettema T.J., Chlebowski A., Dzionek K., Nijtmans L.G., Huynen M.A., Holt I.J. TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res. 2011;39:4284–4299. doi: 10.1093/nar/gkq1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.B., Asin-Cayuela J., Camara Y., Shi Y., Pellegrini M., Gaspari M., Wibom R., Hultenby K., Erdjument-Bromage H., Tempst P., Falkenberg M., Gustafsson C.M., Larsson N.-G. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- Peralta S., Wang X., Moraes C.T. Mitochondrial transcription: lessons from mouse models. Biochim. Biophys. Acta. 2011 doi: 10.1016/j.bbagrm.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remaut H., Waksman G. Protein–protein interaction through beta-strand addition. Trends Biochem. Sci. 2006;31:436–444. doi: 10.1016/j.tibs.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Ringel R., Sologub M., Morozov Y.I., Litonin D., Cramer P., Temiakov D. Structure of human mitochondrial RNA polymerase. Nature. 2011;478:269–273. doi: 10.1038/nature10435. [DOI] [PubMed] [Google Scholar]
- Roberti M., Loguercio Polosa P., Bruni F., Musicco C., Gadaleta M.N., Cantatore P. DmTTF, a novel mitochondrial transcription termination factor that recognises two sequences of Drosophila melanogaster mitochondrial DNA. Nucleic Acids Res. 2003;31:1597–1604. doi: 10.1093/nar/gkg272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberti M., Fernandez-Silva P., Loguercio Polosa P., Fernandez-Vizarra E., Bruni F., Deceglie S., Montoya J., Gadaleta M.N., Cantatore P. In vitro transcription termination activity of the Drosophila mitochondrial DNA-binding protein DmTTF. Biochem. Biophys. Res. Commun. 2005;331:357–362. doi: 10.1016/j.bbrc.2005.03.173. [DOI] [PubMed] [Google Scholar]
- Roberti M., Bruni F., Loguercio Polosa P., Gadaleta M.N., Cantatore P. The Drosophila termination factor DmTTF regulates in vivo mitochondrial transcription. Nucleic Acids Res. 2006;34:2109–2116. doi: 10.1093/nar/gkl181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberti M., Loguercio Polosa P., Bruni F., Manzari C., Deceglie S., Gadaleta M.N., Cantatore P. The MTERF family proteins: mitochondrial transcription regulators and beyond. Biochim. Biophys. Acta. 2009;1787:303–311. doi: 10.1016/j.bbabio.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Roberti M., Loguercio Polosa P., Bruni F., Deceglie S., Gadaleta M.N., Cantatore P. MTERF factors: a multifunction protein family. Biomol. Conc. 2010;1:215–224. doi: 10.1515/bmc.2010.015. [DOI] [PubMed] [Google Scholar]
- Ruiz De Mena I., Lefai E., Garesse R., Kaguni L.S. Regulation of mitochondrial single-stranded DNA-binding protein gene expression links nuclear and mitochondrial DNA replication in Drosophila. J. Biol. Chem. 2000;275:13628–13636. doi: 10.1074/jbc.275.18.13628. [DOI] [PubMed] [Google Scholar]
- Shutt T.E., Shadel G.S. A compendium of human mitochondrial gene expression machinery with links to disease. Environ. Mol. Mutagen. 2010;51:360–379. doi: 10.1002/em.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spåhr H., Samuelsson T., Hällberg B.M., Gustafsson C.M. Structure of mitochondrial transcription termination factor 3 reveals a novel nucleic acid-binding domain. Biochem. Biophys. Res. Commun. 2010;397:386–390. doi: 10.1016/j.bbrc.2010.04.130. [DOI] [PubMed] [Google Scholar]
- Takata K., Yoshida H., Hirose F., Yamaguchi M., Kai M., Oshige M., Sakimoto I., Koiwai O., Sakaguchi K. Drosophila mitochondrial transcription factor A: characterization of its cDNA and expression pattern during development. Biochem. Biophys. Res. Commun. 2001;287:474–483. doi: 10.1006/bbrc.2001.5528. [DOI] [PubMed] [Google Scholar]
- Wenz T., Luca C., Torraco A., Moraes C.T. mTERF2 regulates oxidative phosphorylation by modulating mtDNA transcription. Cell Metab. 2009;9:499–511. doi: 10.1016/j.cmet.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yakubovskaya E., Mejia E., Byrnes J., Hambardjieva E., Garcia-Diaz M. Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription. Cell. 2010;141:982–993. doi: 10.1016/j.cell.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials.