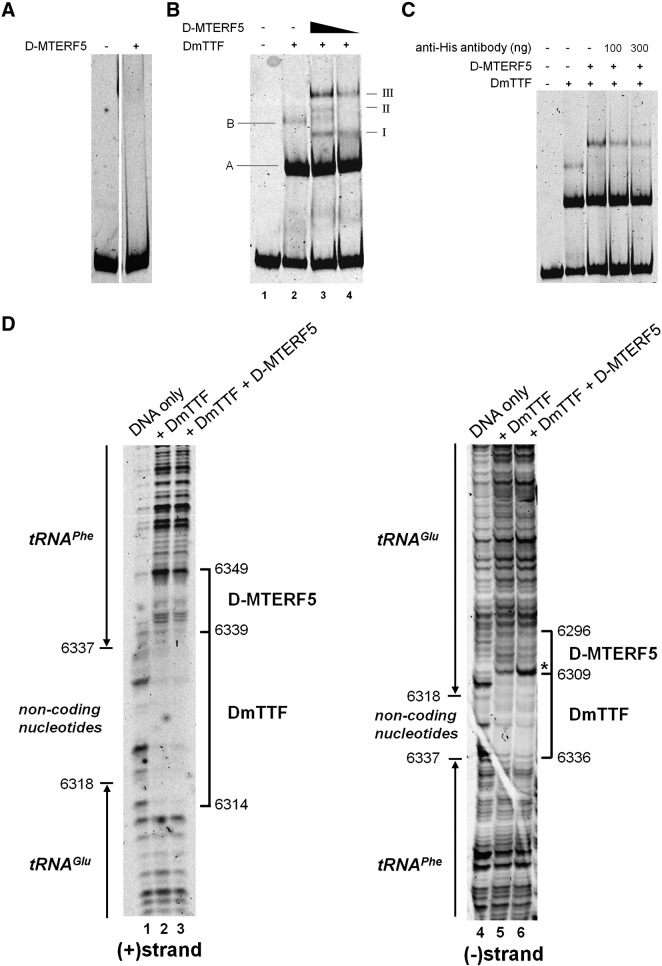
Fig. 3.
Analysis of the DNA-binding properties of D-MTERF5. EMSA and footprinting assays were employed to test the interaction of D-MTERF5 with mtDNA at the tRNAGlu/tRNAPhe boundary region containing the T1 DmTTF binding site. A DNA fragment spanning position nt 6261–6399 of D. melanogaster mtDNA (GenBank ID: NC_001709) labeled with cyanine5 at the 5′-ends was used as a probe in the binding reactions. (A) EMSA was conducted by incubating the DNA probe (25 fmol) with 50 ng (about 0.8 pmol) of recombinant purified D-MTERF5. (B) EMSA was conducted by incubating the DNA probe (25 fmol) with 25 or 12.5 ng (about 0.4 and 0.2 pmol, respectively) of recombinant purified D-MTERF5 in the presence of 50 ng (about 1.2 pmol) of recombinant purified DmTTF. Complexes formed by DmTTF are indicated by capital letters, and complexes obtained following the addition of D-MTERF5 are indicated by Roman numerals. (C) The indicated amounts of anti-6xHis antibodies were added to the EMSA reaction mixture containing 25 fmol of DNA and 50 ng of both D-MTERF5 and DmTTF (about 0.8 and 1.2 pmol, respectively). (D) DNase I protection analysis was conducted on protein–DNA complexes obtained by adding to 100 fmol of DNA fragment labeled at the 5′-end of either (+) or (−) strand, 500 ng (about 8.2 pmol) of purified DmTTF or 500 ng of DmTTF (about 8.2 pmol) plus 250 ng of D-MTERF5 (about 4.0 pmol). Protected regions are indicated by brackets; the asterisk indicates the DNase I-hypersensitive site.