Abstract
The circadian clock is a highly conserved timing system, resonating physiological processes to 24-hour environmental cycles. Circadian misalignment is emerging as a risk factor of metabolic disease. The molecular clock resides in all metabolic tissues, the dysfunction of which is associated with perturbed energy metabolism. In this article, we will review current knowledge about molecular mechanisms of the circadian clock and the role of clocks in the physiology and pathophysiology of metabolic tissues.
Keywords: circadian clocks, metabolism, metabolic disease
Introduction
The circadian (from Latin: circa, about; diem, day) clock is a highly conserved timing system, resonating physiological processes to 24-hour environmental cycles [1]. Daily rhythms of natural light, environment temperature, and food availability set the pace of circadian clocks, which run in almost all mammalian cells [2] (Figure 1). Chronic misalignment between internal clocks and environmental rhythms, including extended work hours, shift work, and frequent time-zone travel, increasingly exaggerate the global pandemic of obesity and metabolic disease [3]. We will review current knowledge on the molecular mechanism of circadian clocks, its role in tissue metabolism, and development of metabolic disease.
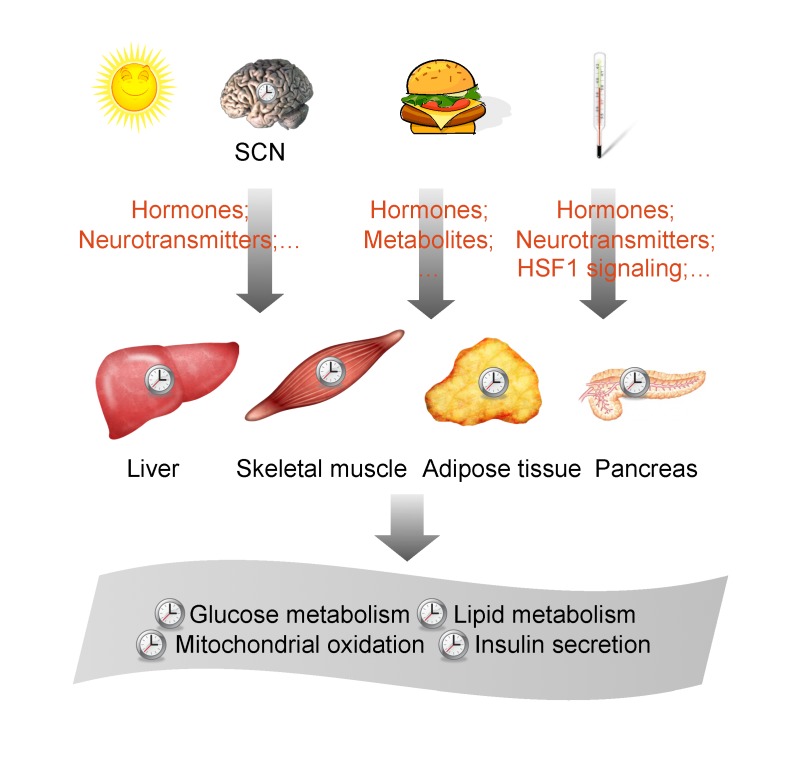
Figure 1.
Architecture of the circadian timing system. Environmental cues, such as light, temperature, and food, reset the body clock through multiple pathways. The suprachiasmatic nucleus (SCN) of the brain is synchronized by light/dark cycles and orchestrates the daily oscillation of internal clocks in different tissues, such as liver, skeletal muscle, adipose tissue, and pancreas, through hormones and neurotransmitters. Environmental temperature cycles reset the body clock through cellular heat shock signaling and humoral/neural pathways. Food availability is also a potent time giver and entrains peripheral clocks through nutrient-sensing and hormonal pathways. The synchronization of different tissue clocks produce coordinated circadian rhythms of metabolic processes, including glucose metabolism, lipid metabolism, mitochondrial oxidation, and insulin secretion.
The Molecular Architecture of Circadian Clocks
The circadian timing system is organized hierarchically in mammals (Figure 1). The suprachiasmatic nucleus (SCN) of the hypothalamus functions as the master pacemaker, synchronizing circadian rhythms of physiological processes in other tissues [4]. However, the molecular architecture of circadian clocks, comprised of clock genes and their regulators, is generally similar among different cells [5]. Generally, the circadian clock has two important features. One is persistence. In constant conditions, the circadian clock keeps ticking. This oscillatory system takes the advantage over a purely driven system, probably by preparing the physiological processes before external changes occur (e.g., sunrise, rise and drop of temperature, and food availability) [6]. The other is entrainment. Organisms never live in a constant environment. Environmental cycles modulate the circadian clock to adapt to a new schedule by varying the phase (the time to show peak). The phase-resetting mechanisms act via changing the endogenous period length or the amplitude in response to a time cue. These features are embedded in the molecular mechanism of the clockwork. Circadian timekeeping occurs at the cellular level by virtue of transcriptional-translational auto-regulatory feedback loops composed of transcription activators BMAL1 and CLOCK and their target genes Period (PER1, PER2, and PER3) and Cryptochrome (CRY1 and CRY2) [1,7]. PERs and CRYs accumulate rhythmically and form an inhibition complex against BMAL1-CLOCK to shut down transcription. The pace of this auto-feedback loop is controlled by various regulatory mechanisms, including post-translational modifications of clock proteins by phosphorylation, acetylation, poly-ADP-ribosylation, and ubiquitination [8-10]. The second loop consists of nuclear receptors REV-ERB (α and β) and ROR (α, β, and γ) [11]. REV-ERBs are transcriptional repressors of BMAL1 gene by competitively binding to BMAL1 promoter against activator RORs [12-15]. The combination of BMAL1-CLOCK-binding elements and REV-ERB/ROR-binding elements along the regulatory region of genes sets the circadian rhythm of gene expression [16]. The physical interaction between PER2 and nuclear receptors contributes to fine-tuning the rhythms. In parallel to the role as a negative regulator of BMAL1/CLOCK-mediated transcription, PER2 acts as a positive regulator of BMAL1 transcription, possibly through binding to REV-ERBα and other nuclear receptors [17,18]. The loop-on-loop architecture of the clock ensures the persistence of the rhythm. Moreover, these molecular oscillators provide the biochemical basis to reset the internal rhythm in response to environmental cycles.
Resetting Mechanisms of Circadian Clocks
Environment cues, such as light, temperature, and food, play an essential role in resetting the pace of circadian clocks through multiple pathways (Figure 1). Daily cycles of natural light and temperature serve as two reliable timing signals for mammals. Transmitted through neural connections from the eye, light entrains the central pacemaker-SCN, through rapid induction of clock genes PER1 and PER2 [19]. In response to light, neurotransmitters glutamate and pituitary adenylate cyclase-activating peptide are released to the SCN neurons and activates cyclic AMP (cAMP) production, calcium signaling, and MAPK cascade, leading to activation of acute-response transcription factor, such as cAMP-response element binding protein (CREB). CREBs drive the immediate induction of PER genes. Actually, rapid induction of PER genes is the principal mechanism to entrain circadian clocks. Studies by Schibler and colleagues demonstrate that multiple pathways of cell signaling, including cAMP, glucocorticoid hormone, protein kinase C, and calcium pathways, synchronize cultured cells through an initial surge of PER expression [20,21]. The signaling between cellular metabolism and the circadian clock is extensive. A recent genome-wide RNAi screen in human cells show that perturbation of components from a variety of cellular processes, such as insulin signaling, hedgehog signaling, cell cycles, and folate metabolism, can affect clock oscillation [22]. Temperature oscillation resets all the body clocks except the SCN due to the cellular network feature of the central pacemaker [23,24]. Recent studies pinpoint circadian protein heat shock transcription factor 1 (HSF1) as a major molecular mediator of the temperature entrainment of peripheral clocks [10,25]. At the organismal level, humoral and neural pathways between the hypothalamic thermal center and the body may participate in the temperature entrainment.
Food availability can also reset peripheral clocks. Limiting the food to the light phase of nocturnal animals can shift circadian clocks in liver and other peripheral organs to the opposing phase [26,27]. Similar to temperature oscillation, restricted feeding schedule has no effect on the SCN clock [26]. The SCN clock and the feeding rhythm both transmit resetting signals to peripheral organs. Glucocorticoid receptor in the liver mediates the SCN-dependent signaling and counteracts the phase deviation from the central pacemaker [28]. Liver-specific glucocorcoid receptor knockout mice exhibit faster adaptation in the liver clock to the new feeding rhythm; whereas, the kidney clock exhibits the indistinguishable adaptation rate as the wildtype. Poly-ADP-ribosyl transferase 1 (PARP1) mediates the feeding-dependent signaling and facilitates the phase shift through poly-ADP-ribosylation of CLOCK and rapid induction of PER2 gene [29]. Food availability might affect circadian clocks through nutrient sensing pathways since it immediately affects nutrient flux into cells [2]. Feeding/fasting modulates cellular NAD+ and AMP levels, which might serve as nutrient sensors [30,31]. Both PARP1 and protein lysine-specific deacetylase SIRT1 utilize NAD+ as the donor substrate. SIRT1 modulates the protein stability of PER2 and the repressor recruitment of BMAL1 through direct deacetylation on these proteins [32-34]. AMP-activated protein kinase (AMPK) phosphorylates and destabilizes CRY1 to regulate circadian clocks, providing another pathway of metabolic regulation [35].
The Metabolic Function of Circadian Clocks in Peripheral Tissues
In mammals, circadian clocks modulate physiological processes by orchestrating daily rhythms of transcriptomes and metabolomes in tissue metabolism [36-38] (Figure 1). The fact that mice with germ-line disruptions of circadian clocks exhibit perturbed glucose homeostasis [39,40] spurred studies on the potential role of this biological timing system in peripheral tissues. Also, variants of clock genes are associated with susceptibility to type 2 diabetes [41-43]. In this section, we will review current knowledge linking tissue circadian clocks to energy homeostasis.
Hypothalamus: Regulation of Energy Balance
In the energy balance equation, energy store is determined by energy intake and energy expenditure, both of which are regulated by the hypothalamus [44]. The hypothalamus is a brain region that integrates nutritional (glucose, amino acids, and lipids) and hormonal (leptin, insulin, ghrelin, and cholecystokinin) signals to modulate energy balance [45]. In particular, the arcuate nucleus, ventromedial, dorsomedial, and lateral hypothalamic nuclei are major nodes in the complex network that regulates energy balance and affects the development of metabolic disease. Circadian clocks in these neural circuits may be involved in the control of energy balance (Table 1). CLOCKΔ19 mice, which are arrhythmic in behavior due to a mutation in the CLOCK gene [46,47], exhibit attenuated rhythms of food intake, contributing to hyperphagy and obesity [39]. Observational studies in diet-induced obese animals show that obesity alters the circadian rhythm of feeding behavior, leading to increased food intake during the rest phase and decreased food consumption in the activity phase [48]. The biochemical mechanisms remain largely unknown. Daily changes in cellular metabolites, such as AMP and NAD+, modulate circadian oscillation [35,49,50]. Nutrient burden might perturb the metabolite rhythms and thus reset the hypothalamic clock.
Table 1. Metabolic phenotypes in mice with mutations in circadian clocks.
| Protein | Mutation | Metabolic Phenotype | Reference |
| CLOCK | Whole-body loss-of-function | Attenutated feeding rhythm, obesity, hyperphagy, hyperlipidemia, hyperglycemia, hepatic steatosis, hypoinsulinemia | [39,111] |
| BMAL1 | Whole-body knockout | Glucose intolerance, hypoinsulinemia, increased respiratory quotient, reduced fat storage, increased circulating fatty acid, increased ectopic fat formation in liver and muscles, hypoinsulinemia | [59,87,103,111] |
| BMAL1 | Liver-specific knockout | Hypoglycemia in the rest phase | [59] |
| CRY1 CRY2 | Whole-body double knockout | Glucose intolerance and constitutively high levels of circulating corticorsterone | [60] |
| RER-ERBα REV-ERBβ | Whole-body double knockout | Hepatic steatosis, hyperglycemia, hyperlipidemia | [11,61] |
| HDAC3 | Liver-specific knockout | Hepatic steatosis | [76] |
| PGC-1α | Whole-body double knockout | Abnormal diurnal rhythms of activity, body temperature and metabolic rate | [78] |
| AMPK | Whole-body knockout in alpha1 subunit | Dampened rhythm in body temperature | [90] |
Limiting food availability promotes physical activity several hours ahead of the mealtime [51,52]. The hypothalamus is also involved in this circadian food anticipation behavior. Lesion studies demonstrate that the SCN clock is not involved in food anticipation and suggest the existence of a food-entrainable oscillator in the hypothalamus. However, the identity of this oscillator is controversial and still undergoes investigation. Knockout studies suggest that the oscillator might function without BMAL1 [53-55].
Liver: Regulation of Glucose, Lipid, and Amino Acid Homeostasis
The liver is a central metabolic organ in the homeostasis of glucose, lipid, and amino acid. Opposing metabolic processes, such as glycolysis/gluconeogenesis, and lipogenesis/fatty acid oxidation, take place in the liver, necessitating the need for temporal separation [56]. Circadian clocks play a significant role in the regulation of hepatic function. Roughly 10 percent of transcripts undergo circadian oscillation in the liver, including enzymes and regulators of major metabolic processes [57,58]. For example, the rate-limiting enzymes of glycolysis and gluconeogenesis oscillate and peak in the early morning and early evening, respectively. Metabolomic studies have shown that a broad variety of hepatic metabolites oscillate in a daily manner [37]. Metabolic phenotyping studies of clock mutant mice illustrate the importance of a function clock on liver metabolism (Table 1). CLOCKΔ19 mice exhibit hyperlipidemia, hyperglycemia, and hepatic steatosis [39]. Liver-specific knockout of BMAL1 results in loss of rhythmic expression of glucose regulatory genes and abnormal glucose homeostasis [59]. Genetic loss of CRY1 and CRY2 leads to glucose intolerance and constitutively high levels of circulating corticorsterone [60]. Liver-specific double knockout of REV-ERBα and β results in hepatic steatosis and impaired rhythmic expression of metabolic genes [11,61].
The circadian regulation of liver metabolism is well demonstrated by studies on the rate-limiting gluconeogenic enzyme, phosphoenolpyruvate carboxylase (PEPCK). The PEPCK activity is diurnal in mouse liver [62], contributing to the diurnal rhythm of hepatic glucose production [63]. The oscillation of enzymatic activity is mainly due to cyclic cellular accumulation of PEPCK, since this enzyme has a short half-life and the major control mechanism is gene expression [64]. The liver clock modulates the rhythmic expression of PEPCK via multiple pathways. CREB and forkhead box protein O1 (FOXO1) are two major transcription factors that drive PEPCK expression [65,66]. Feeding/fasting cycles generate daily oscillation of metabolic hormones, such as glucagon, glucocorticoid hormone, thyroid hormone, and insulin. Glucagon activates CREB through the cAMP/PKA axis. Kay and colleagues have shown that the circadian repressor CRYs modulate the daily strength of the glucagon signaling by affecting cellular accumulation of cAMP [67]. Glucocorticoid hormone is released in a diurnal manner and activates the transcription of PEPCK [28,64,68]. Recently, Evans and colleagues have characterized the glucocorticoid-dependent interaction between CRYs and glucocorticoid receptor [60]. CRYs repress both glucagon and glucocorticoid hormone signaling and thus constitute a negative circadian pathway to the transcriptional control of PEPCK. The REV-ERB/ROR pair directly regulates PEPCK expression [69]. PPARgamma coactivator-1α (PGC-1α) exhibits circadian pattern of gene expression and contributes to PEPCK gene by physical interaction with FOXO1 and ROR [70]. In summary, the circadian regulation of PEPCK activity is through coordinated actions of systemic hormonal cues and local clockwork (Figure 2). Recent studies showed that shift work desynchronizes the body clock from environmental rhythms in part by perturbing the diurnal rhythm of PEPCK activity, leading to impaired gluconeogenic potential [71].
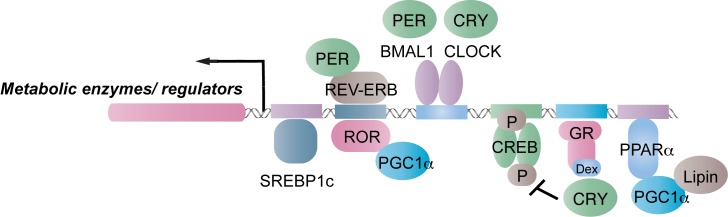
Figure 2.
Molecular mechanisms of circadian expression of metabolic enzymes/regulators. The circadian clock regulates tissue metabolism mostly through expression of metabolic enzymes and regulators. Transcription factors of the circadian clock, such as REV-ERB, ROR and BMAL1/CLOCK can directly participate in the transcriptional control. Nutritional and hormonal signaling pathways, such as SREBP1c, cAMP/CREB, GRα, PPARα/PGC-1α/Lipin, are metabolic regulators. SREBP1c, glucocorticoid signaling, and the PPARα/PGC-1α/Lipin axis are under circadian control. CRY interacts with cAMP/CREB and GRα signaling to transmit temporal signals. In addition, the PER/REV-ERB axis and PGC-1α/ROR axis fine-tune the transcriptional network.
Another important facet of liver function is lipid metabolism. Perturbation of lipid homeostasis in the liver is involved in the development of non-alcoholic fatty liver disease and diabetes [72]. In recent years, molecular mechanisms of how circadian clocks regulate lipid metabolism have been studied extensively (Figure 2). It is been known for years that regulators of lipid metabolism oscillate in the liver. These regulators are directly or indirectly regulated by circadian clocks. REV-ERB proteins represent the direct output pathway of the circadian clock. Genetic studies demonstrate that REV-ERBα regulates the circadian oscillation of sterol element binding protein 1c (SREBP1c) and thus the hepatic and plasma levels of triglyceride, cholesterol, and lipoproteins [73]. REV-ERB recruits nuclear receptor corepressor (NCoR) and histone deacetylase 3 (HDAC3) to repress the transcription of metabolic genes involved in lipid metabolism [74]. The dysfunction of the REV-ERB/NCoR/HDAC3 complex leads to hepatic steatosis and hyperlipidemia [11,61,75,76]. The importance of the circadian regulation is evident from the observation that most of the lipid metabolic genes have no difference in the daily average abundance between control and transgenic mice, but have significant perturbed diurnal expression patterns. Feeding rhythm remains unchanged in REV-ERB deficient mice, indicating REV-ERB regulates lipid metabolism independent of systemic cues [73]. Other master regulators can be activated by feeding rhythm and thus are subject to the SCN clock. Yang and coworkers observed the diurnal cycling of all PPAR family members in mouse liver [77]. PPARα, the dominant isoform in liver, promotes fatty acid oxidation and reaches its peak around early evening. The PPARgamma coactivator, PGC-1α, oscillates in phase with PPARα [78]. The amplifier of the PPARα/PGC-1α module, lipin 1, oscillates in phase with both PPARα and PGC-1α [58,79]. The concerted oscillation of the PPARα/PGC-1α/lipin1 regulatory module promotes the utilization of fatty acids at the beginning of the nighttime feeding phase, preparing the hepatocyte to meet the increased energy consumption from physical activities. Transcriptional regulator of the biosynthetic arm of lipid metabolism is also cycling. SREBP1c is expressed in a diurnal manner in the liver and orchestrates the biosynthesis of fatty acid and triglyceride [80,81]. Both the PPARα/PGC-1α/lipin1 module and the SREBP1c module are not only driven by systemic cues, but also modulated by the liver clock. The physical interaction between PPARα and PER2 and the REV-ERB/Insig2/SREBP1c axis may contribute to the crosstalk between the lipid metabolic network and the liver clock [18,73].
Adipose Tissue: Regulation of Fat Mass and Body Temperature
Adipose tissue is a fat depot and a major endocrine organ. About 80 percent of fat is stored in adipose tissue and the rest resides in liver and muscle. Failure to absorb lipids and fatty acids in adipose tissue leads to ectopic accumulation of fat in liver and muscles. In recent years, adipose tissue has been recognized as an important metabolic endocrine organ. It synthesizes and releases hormones, including leptin, adiponectin, and tumor necrosis factor-α, collectively known as adipokines. In rodent and human studies, diurnal expression profiles of clock and clock-controlled genes have been characterized in adipose tissue, indicating the presence of a functional clock [82-85]. Circadian oscillations in plasma leptin, glucose, triglycerides, free fatty acids, and LDL cholesterol were reported [85], which is likely under the control of the adipocyte clock.
Circadian rhythms in adipose tissue maintain the tissue and energy homeostasis (Table 1). Shimba and colleagues reported that BMAL1 is an essential regulator of adipogenesis and lipid metabolism in matured adipocytes [86]. Embryonic fibroblast cells deficient in BMAL1 gene fail to differentiate into adipocytes except introduction of exogenous BMAL1 copy. Bmal1 knockout mice have increased respiratory quotient, reduced fat storage, increased circulating fatty acid, and increased ectopic fat formation in liver and muscles [87]. Most of the genes involved in adipocyte function (e.g., PPARγ2, C/EBPα, SREBP1c, and lipin1) are expressed at a low level in BMAL1 deficient mice, whereas the preadipocyte marker PREF-1 is expressed at a high level. The defective adipogenesis in BMAL1 knockout mice results in a failure for adipose tissue to expand upon nutrient excess, and thus ectopic accumulation of fat in liver and muscles. Intriguingly, the differences in adipose tissue size appear in adults but not juveniles of BMAL1 knockout mice [88], linking a functional clock to longevity. In parallel to direct regulation by clock genes, rhythmic expression of a majority of nuclear receptors in adipose tissue might exert a large-scale coordination of signaling pathways to regulate adipocyte biology [77,89]. PPARγ, a target of anti-diabetic thiazolidinediones, is a master regulator of adipogenesis and adipocytic energy metabolism [90,91]. The oscillatory cellular accumulation of PPARγ might contribute to the temporal fluctuation of metabolism in adipose tissue [77]. REV-ERBα modulates adipogenesis and PPARγ induction [92]. A recent study shows that systemic administration of a REV-ERBα agonist in diet-induced obese mice promotes leanness by reducing fat mass and improving dyslipideamia and hyperglycemia [93]. Expression of an array of metabolic genes in adipose tissue is affected by REV-ERBα agonist treatment. This is the first study to demonstrate that a drug targeted on a circadian regulator can improve syndromes of metabolic disease. In addition, adipose tissue participates in temperature homeostasis by thermogenesis. It has been known for decades that body temperature follows a circadian rhythm [94]. Under constant environmental conditions, body temperature is higher in the activity phase and lower in the rest phase. Circadian clocks play an essential role in orchestrating temporal fluctuation of body temperature. Mice with disruption in both CRY genes exhibit arrhythmic pattern of daily body temperature and heat production [95]. Though studies with adipose tissue-specific clock mutant mice are not available, knockout studies with different isoforms of AMPK catalytic subunit provide an important clue [96]. AMPK has two catalytic subunits: α1 and α2. The circadian expression patterns of core clock genes are severely affected in adipose tissue of AMPKα1-/-, in line with a dampened rhythm in body temperature. In contrast, AMPKα2-/- mice exhibit normal rhythms in core clock gene expression in adipose tissue and a normal circadian rhythm in body temperature.
Skeletal Muscle: Regulation of Oxidative Metabolism and Muscle Mass
Skeletal muscle is a major organ for glucose disposal in response to food intake and insulin [72]. Impairment of glucose uptake in skeletal muscle contributes to development of type 2 diabetes. About 3.4 percent of transcripts oscillate in a circadian manner in skeletal muscle, and as expected, the largest cluster (18 percent) of circadian transcripts represents genes involved in intermediary metabolism [97,98]. Interestingly, the vast majority of these circadian metabolic transcripts are involved in lipid homeostasis, including nuclear receptors and their co-regulators [98]. As an energy-demanding organ, skeletal muscle relies heavily on fatty acids as a fuel source. Nuclear receptors RORα and REV-ERBβ are master regulators of lipid homeostasis, independent of their circadian function [99,100]. PPARgamma coactivator-1β (PGC-1β) acts as a ligand of nuclear receptor ERR (estrogen-related receptor) and activates expression of medium chain acyl CoA dehydrogenase (Mcad) [101]. PGC-1α is also a circadian gene in skeletal muscle, orchestrating transcriptional program of oxidative metabolism, skeletal myofiber switching, and circadian rhythms [70,102]. The circadian rhythms of these metabolic regulators contribute to the maintenance of mitochondrial mass and function in skeletal muscle (Table 1). Mice with disrupted circadian clocks exhibit abnormal mitochondrial morphology and cellular respiration in muscles [103], in line with a decreased metabolic rate [87]. MyoD, a master regulator of muscle physiology, is expressed in a circadian manner under the direct control of BMAL1-CLOCK, and this regulation is involved in the homeostasis of muscle mass and function [103]. However, whether disruption of circadian clocks in skeletal muscle contributes to the etiology of metabolic disease requires future investigation. It is possible that anti-diabetics can improve glucose homeostasis by restoring circadian rhythms in skeletal muscle. A recent report suggests that Ramipril, an anti-diabetic by inhibiting angiotensin-converting enzyme, can dramatically induce expression of core clock genes in skeletal muscle [104]. More than a third of the circadian transcriptome reach their peak expression in the middle of the activity phase, when animals are most physically active and feeding [98]. Though mechanisms remain elusive, rhythmic behavior, such as feeding, might reset the circadian clock in skeletal muscle through changes in cellular redox ratio and nutrient flux [56].
Pancreas: Regulation of Insulin Secretion, Beta-Cell Mass, and Glucose Homeostasis
Pancreatic islet beta-cells secrete insulin in response to nutrients and hormones and play an essential role in glucose homeostasis. In mammals, deterioration in beta-cell function and partial loss of beta-cell mass trigger the transition from an obese, insulin-resistant state to a full-blown type 2 diabetes [105]. Early studies by Polonsky and colleagues show that rhythmic control of insulin secretion is disturbed in patients with type 2 diabetes, whereas the obese retains a largely normal pattern of insulin release [106,107], suggesting that disturbed rhythms of insulin secretion contributes to etiology of type 2 diabetes.
Several studies have demonstrated the presence of an autonomous circadian clock in beta-cells. Peschke E. and Peschke D. reported the circadian pattern of insulin release in isolated rat islets [108]. Major clock genes and clock-controlled genes, including glucose transporter 2 and glucokinase, are expressed in a rhythmic manner in pancreas [109,110]. Real-time imaging studies of tissue explants show that promoter activities of PER2 and BMAL1 genes exhibit circadian rhythms in islets and can be abolished in clock mutants, identifying the presence of autonomous circadian clocks in beta-cells [111,112].
The dysfunction of the pancreas clock affects insulin secretion and glucose homeostasis (Table 1). Islet-specific disruption of circadian clocks in mice causes impaired insulin secretion and reduced circulating levels, accounting for the diabetic phenotype-hyperglycemia and impaired glucose tolerance [111,112]. Both clock mutant mice CLOCKΔ19 and BMAL1-/- exhibit impaired glucose tolerance, which is associated with hypoinsulinaemia, reduced nutrient/hormone-induced insulin secretion, and mild defects in islet size and proliferation [111]. The architecture of pancreas remains intact, suggesting that the major defect in clock mutant islets is dysregulated beta-cell function. Consistent with these phenotypes, microarray analysis of islet samples show decreased levels in clock genes, significant alterations in vesicle-docking and trafficking factors, and increased levels in apoptosis factors [111]. Meanwhile, circadian expression patterns of key genes involved in glucose metabolism, insulin signaling, cell cycle, and beta-cell growth are altered in clock mutant islets. The fact that perturbations of genes involved in different feedback loops of circadian clocks causes impaired beta-cell function and cell mass [111-113] highlights the important role of circadian clocks in the pancreas biology.
Circadian Misalignment Contributes to Metabolic Dysfunction
The growing knowledge about the circadian regulation of tissue metabolism illuminates the idea that the alignment between the internal clock and environmental cycles is important for the body’s health. In modern society, changes in lifestyles perturb the alignment in several ways and have broad implications on metabolic dysfunction [114]. Jet lag represents the rapid misalignment. Jet lag is a consequence of crossing time zones too rapidly for the internal clock to adjust to local environmental cycles, including light and food. Despite a large number of studies on jet lag and sleep disorder [115], how jet lag impacts metabolism is unknown. Shift work represents the chronic misalignment. About 8.6 million Americans perform shift work [116]. Work in the night/early morning resets the internal rhythm against the environmental cycles. It has been known that shift work is strongly associated with obesity and metabolic disease [117]. Recent studies show that shift work mimetic perturbs the circadian clock and energy metabolism, including glucose production and glycogen/lipid contents, in mouse liver [71]. Interestingly, limiting food access to the dark phase can effectively rescue the clock and metabolic dysfunction caused by shift work. It is evident that alignment of the feeding rhythm to the light/dark cycle counteracts the metabolic dysfunction caused by shift work.
Circadian Dlock-Targeted and -Guided Treatment of Metabolic Disease
Our understanding of circadian clock architecture, coordination, and physiology has grown rapidly in the past few decades. In parallel, the daily rhythms of physiological processes are emerging as an important factor to improve therapeutic outcomes of metabolic disease. Studies on the synthetic ligands of REV-ERBα illuminate the idea that direct manipulation of the circadian rhythms may have beneficial metabolic outcomes [93]. Besides direct pharmaceutical interference, restoration of the normal circadian rhythms by light exposure, food regimens, temperature oscillation, and scheduled exercise is also promising. Food regimens provide a time cue for the internal clock and have been demonstrated to improve energy homeostasis in obese animals. Hatori and colleagues have reported that restricted feeding with isocaleric food can protect against obesity and hepatic steatosis in diet-induced obese mice [31]. Restricted feeding improves the metabolic function, probably through major nutrient sensing pathways, such as CREB, mTOR, and AMPK, as well as the daily oscillation of circadian clocks in these animals. Calorie restriction is well known to increase lifespan from worms to mammals [118]. Studies have shown that calorie restriction can affect circadian timing in the SCN clock [119] and is likely to modulate peripheral clocks through feeding rhythm. Another food regimen, intermittent fasting, has been proposed to improve energy metabolism through rectifying clock oscillation [120]. Scheduled exercise has pronounced effects on improving insulin sensitivity [121] and can shift the phase of clock oscillation in the skeletal muscle [122]. Key metabolic sensors, such as PGC-1α, AMPK, SIRTs and mTOR, are likely the molecular underpinnings of these potential therapies. Moreover, knowledge of circadian rhythm of different physiological processes could aid in the improvement of drug efficacy by timed delivery. Recently, Hermida and colleagues have shown that compared with all treatments upon waking, blood pressure-lowering treatment at bedtime has significantly improved cardiovascular conditions in diabetic patients [123].
Conclusions and Outlook
The general feature of the circadian clock is shared across all organisms, from single-cellular cyanobacteria to multi-cellular plants and mammals. This rare commonness pronounces the fundamental importance of circadian rhythms. Since the first genetic experiment by Bünning in 1935, our knowledge of the organization, molecular nature, and synchronization mechanisms of circadian clocks have expanded dramatically. In the past decade, dissecting the circadian regulation of metabolic systems has revealed the essential role of internal clocks in energy homeostasis, ushering in a “timed” era to explore chronotherapeutics of metabolic disease. However, most of the previous studies using genetic mutants cannot distinguish between effects of impaired circadian rhythms (clock function) and those of perturbed basal activity/expression (non-clock function) of target genes on metabolism. Future studies require more rhythm-targeted approaches to identify the physiological significance of circadian oscillation. For example, timed manipulation of clock components or signaling (i.e., induction/inhibition of a clock gene at a specific time window or knock-in animal models) would flatten or reverse the rhythms of clock-controlled genes and thus can specifically assay the effects of perturbed rhythm.
Despite the technical challenges, several important questions remain to be addressed. First, what are the molecular and cellular output pathways of peripheral clocks in the regulation of local biological processes? Accumulating evidence has linked peripheral clocks to many local biological processes. Large-scale high throughout assays would produce detailed molecular features [124], generating new hypotheses of output pathways. For example, deep sequencing in fruit flies uncovered extensive control of non-coding RNA expression, alternative splicing, and RNA editing, which likely contributes to tissue specificity of output pathways [125]. Second, how does food availability or circadian disruption at the systemic or the local levels affect circadian clocks? Scheduled feeding can effectively reset the circadian clock through nutritional and hormonal pathways. However, the molecular signaling and cellular communication pathways are largely uncharacterized. Chronic circadian disruption by shift work is emerging as a risk factor for metabolic disease. Delineation of the molecular, cellular, and systemic pathways would enrich the repertoire of therapeutic targets. After decades of extensive studies, knowledge about circadian rhythms is starting to transform our views about the human physiology of metabolism and would bring fundamental benefits to human health in the 21st century.
Acknowledgments
We thank Dr. Xiaoyong Yang and other Yang laboratory members for kind support. M.L. is grateful to Yao Wu for inspiring discussions. This work was supported by fellowship from the China Scholarship Council-Yale World Scholars in the Biomedical Sciences and the Society for Research on Biological Rhythms (SRBR) Research Excellence Award to M.L. M.L. and C.L. contributed equally to this work.
Abbreviations
- AMPK
AMP-activated protein kinase
- BMAL1
brain and muscle Arnt-like protein 1
- CLOCK
circadian locomotor output cycles kaput
- CRY
cryptochrome
- MAPK
mitogen-activated protein kinase
- MyoD
myogenic differentiation
- PARP1
poly-ADP-ribosyl-transferase 1
- PEPCK
peroxisome-proliferator activated receptor
- PER
period
- PGC-1
PPARgamma coactivator
- PPAR
peroxisome-proliferator activated receptor
- REV-ERB
reverse ErbA
- ROR
RAR-related orphan recepto
- SCN
suprachiasmatic nucleus
- SIRT1
Sirtuin1
References
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418(6901):935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330(6009):1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton OM, Cain SW, O’Connor SP, Porter JH, Duffy JF, Wang W. et al. Adverse metabolic consequences in humans of prolonged sleep restriction combined with circadian disruption. Sci Transl Med. 2012;4(129):129ra43. doi: 10.1126/scitranslmed.3003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Saini C, Suter DM, Liani A, Gos P, Schibler U. The mammalian circadian timing system: Synchronization of peripheral clocks. Cold Spring Harb Symp Quant Biol. 2011;76:39–47. doi: 10.1101/sqb.2011.76.010918. [DOI] [PubMed] [Google Scholar]
- Roenneberg T, Merrow M. Life before the clock: Modeling circadian evolution. J Biol Rhythms. 2002;17(6):495–505. doi: 10.1177/0748730402238231. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Reddy AB, Maywood ES. A clockwork web: Circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4(8):649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- Mehra A, Baker CL, Loros JJ, Dunlap JC. Post-translational modifications in circadian rhythms. Trends Biochem Sci. 2009;34(10):483–490. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguay D, Cermakian N. The crosstalk between physiology and circadian clock proteins. Chronobiol Int. 2009;26(8):1479–1513. doi: 10.3109/07420520903497575. [DOI] [PubMed] [Google Scholar]
- Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13(2):125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT. et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485(7396):123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U. et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12(5):441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T. et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418(6897):534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15 Spec No 2:R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y. et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37(2):187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B. et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288(5468):1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24(4):345–357. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Fukada Y. Resetting mechanism of central and peripheral circadian clocks in mammals. Zoolog Sci. 2004;21(4):359–368. doi: 10.2108/zsj.21.359. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93(6):929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured rat-1 fibroblasts. Curr Biol. 2000;10(20):1291–1294. doi: 10.1016/s0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY. et al. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139(1):199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol. 2001;12(18):1574–1583. doi: 10.1016/s0960-9822(02)01145-4. [DOI] [PubMed] [Google Scholar]
- Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330(6002):379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru T, Hattori M, Honda K, Benjamin I, Ozawa T, Takamatsu K. Synchronization of circadian Per2 rhythms and HSF1-BMAL1:CLOCK interaction in mouse fibroblasts after short-term heat shock pulse. PLoS One. 2011;6(9):e24521. doi: 10.1371/journal.pone.0024521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14(23):2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291(5503):490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- Le Minh N, Damiola F, Tronche F, Schutz G, Schibler U. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J. 2001;20(24):7128–7136. doi: 10.1093/emboj/20.24.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142(6):943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Bass J. Circadian clocks in fuel harvesting and energy homeostasis. Cold Spring Harb Symp Quant Biol. 2011;76:63–72. doi: 10.1101/sqb.2011.76.010546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori M, Vollmers C, Zarrinpar A, Ditacchio L, Bushong EA, Gill S. et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15(6):848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y. et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450(7172):1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D. et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F. et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134(2):317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF. et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326(5951):437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134(5):728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, Mohney RP, Vignola KS, Baldi P, Sassone-Corsi P. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci USA. 2012;109(14):5541–5546. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci USA. 2012;109(7):2625–2629. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E. et al. Obesity and metabolic syndrome in circadian clock mutant mice. Science. 2005;308(5724):1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB. et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2(11):e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woon PY, Kaisaki PJ, Braganca J, Bihoreau MT, Levy JC, Farrall M. et al. Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc Natl Acad Sci USA. 2007;104(36):14412–14417. doi: 10.1073/pnas.0703247104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU. et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly MA, Rees SD, Hydrie MZ, Shera AS, Bellary S, O’Hare JP. et al. DIAGRAM consortium, SAT2D consortium. Circadian gene variants and susceptibility to type 2 diabetes: A pilot study. PLoS One. 2012;7(4):e32670. doi: 10.1371/journal.pone.0032670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD. Leptin and beyond: An odyssey to the central control of body weight. Yale J Biol Med. 2011;84(1):1–7. [PMC free article] [PubMed] [Google Scholar]
- Dietrich MO, Horvath TL. Feeding signals and brain circuitry. Eur J Neurosci. 2009;30(9):1688–1696. doi: 10.1111/j.1460-9568.2009.06963.x. [DOI] [PubMed] [Google Scholar]
- Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD. et al. Mutagenesis and mapping of a mouse gene, clock, essential for circadian behavior. Science. 1994;264(5159):719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP. et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89(4):641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y. et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6(5):414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324(5927):654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B. et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324(5927):651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistlberger RE. Neurobiology of food anticipatory circadian rhythms. Physiol Behav. 2011;104(4):535–545. doi: 10.1016/j.physbeh.2011.04.015. [DOI] [PubMed] [Google Scholar]
- Verwey M, Amir S. Food-entrainable circadian oscillators in the brain. Eur J Neurosci. 2009;30(9):1650–1657. doi: 10.1111/j.1460-9568.2009.06960.x. [DOI] [PubMed] [Google Scholar]
- Pendergast JS, Nakamura W, Friday RC, Hatanaka F, Takumi T, Yamazaki S. Robust food anticipatory activity in BMAL1-deficient mice. PLoS One. 2009;4(3):e4860. doi: 10.1371/journal.pone.0004860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistlberger RE, Buijs RM, Challet E, Escobar C, Landry GJ, Kalsbeek A. et al. Food anticipation in Bmal1-/- and AAV-Bmal1 rescued mice: A reply to fuller et al. J Circadian Rhythms. 2009;7:11. doi: 10.1186/1740-3391-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistlberger RE, Yamazaki S, Pendergast JS, Landry GJ, Takumi T, Nakamura W. Comment on “differential rescue of light- and food-entrainable circadian rhythms.”. Science. 2008;322(5902):675. doi: 10.1126/science.1161284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter J, Reick M, McKnight SL. Metabolism and the control of circadian rhythms. Annu Rev Biochem. 2002;71:307–331. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH. et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417(6884):78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M. et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105(39):15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Papp SJ, Yu RT, Barish GD, Uhlenhaut NH, Jonker JW. et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480(7378):552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F. et al. Rev-erbalpha and rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26(7):657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips LJ, Berry LJ. Circadian rhythm of mouse liver phosphoenolpyruvate carboxykinase. Am J Physiol. 1970;218(5):1440–1444. doi: 10.1152/ajplegacy.1970.218.5.1440. [DOI] [PubMed] [Google Scholar]
- Kida K, Nishio T, Yokozawa T, Nagai K, Matsuda H, Nakagawa H. The circadian change of gluconeogenesis in the liver in vivo in fed rats. J Biochem. 1980;88(4):1009–1013. doi: 10.1093/oxfordjournals.jbchem.a133051. [DOI] [PubMed] [Google Scholar]
- Chakravarty K, Cassuto H, Reshef L, Hanson RW. Factors that control the tissue-specific transcription of the gene for phosphoenolpyruvate carboxykinase-C. Crit Rev Biochem Mol Biol. 2005;40(3):129–154. doi: 10.1080/10409230590935479. [DOI] [PubMed] [Google Scholar]
- Gross DN, van den Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27(16):2320–2336. doi: 10.1038/onc.2008.25. [DOI] [PubMed] [Google Scholar]
- Altarejos JY, Montminy M. CREB and the CRTC co-activators: Sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12(3):141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T. et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16(10):1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips LJ, Berry LJ. Hormonal control of mouse liver phosphoenolpyruvate carboxykinase rhythm. Am J Physiol. 1970;219(3):697–701. doi: 10.1152/ajplegacy.1970.219.3.697. [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA. et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318(5857):1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Liu C, Lin JD. PGC-1 coactivators in the control of energy metabolism. Acta Biochim Biophys Sin (Shanghai) 2011;43(4):248–257. doi: 10.1093/abbs/gmr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay JL, Husse J, Bode B, Naujokat N, Meyer-Kovac J, Schmid SM. et al. Circadian desynchrony promotes metabolic disruption in a mouse model of shiftwork. PLoS One. 2012;7(5):e37150. doi: 10.1371/journal.pone.0037150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel VT, Shulman GI. Mechanisms for insulin resistance: Common threads and missing links. Cell. 2012;148(5):852–871. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, Sasso GL. et al. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7(9):e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Lazar MA. The orphan nuclear receptor rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19(6):1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J. et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456(7224):997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T. et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331(6022):1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M. et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126(4):801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447(7143):477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- Finck BN, Gropler MC, Chen Z, Leone TC, Croce MA, Harris TE. et al. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 2006;4(3):199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109(9):1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer M, Lange D, Baler R, Anzulovich A. SREBP-1 as a transcriptional integrator of circadian and nutritional cues in the liver. J Biol Rhythms. 2005;20(3):195–205. doi: 10.1177/0748730405275952. [DOI] [PubMed] [Google Scholar]
- Ando H, Yanagihara H, Hayashi Y, Obi Y, Tsuruoka S, Takamura T. et al. Rhythmic messenger ribonucleic acid expression of clock genes and adipocytokines in mouse visceral adipose tissue. Endocrinology. 2005;146(12):5631–5636. doi: 10.1210/en.2005-0771. [DOI] [PubMed] [Google Scholar]
- Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G. et al. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55(4):962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- Wu X, Xie H, Yu G, Hebert T, Goh BC, Smith SR. et al. Expression profile of mRNAs encoding core circadian regulatory proteins in human subcutaneous adipose tissue: Correlation with age and body mass index. Int J Obes (Lond) 2009;33(9):971–977. doi: 10.1038/ijo.2009.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran S, Xue B, Jusko WJ, Dubois DC, Almon RR. Circadian variations in gene expression in rat abdominal adipose tissue and relationship to physiology. Physiol Genomics. 2010;42A(2):141–152. doi: 10.1152/physiolgenomics.00106.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, Hayashi M. et al. Brain and muscle arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci USA. 2005;102(34):12071–12076. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimba S, Ogawa T, Hitosugi S, Ichihashi Y, Nakadaira Y, Kobayashi M. et al. Deficient of a clock gene, brain and muscle arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PLoS One. 2011;6(9):e25231. doi: 10.1371/journal.pone.0025231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20(14):1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. A wheel of time: The circadian clock, nuclear receptors, and physiology. Genes Dev. 2010;24(8):741–747. doi: 10.1101/gad.1920710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Barak Y, Hevener A, Olson P, Liao D, Le J. et al. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci USA. 2003;100(26):15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JR, Barrick C, Kim KA, Lindner J, Blondeau B, Fujimoto Y. et al. Deletion of PPARgamma in adipose tissues of mice protects against high fat diet-induced obesity and insulin resistance. Proc Natl Acad Sci USA. 2005;102(17):6207–6212. doi: 10.1073/pnas.0306743102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristancho AG, Lazar MA. Forming functional fat: A growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol. 2011;12(11):722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T. et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485(7396):62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschoff J. Circadian control of body temperature. J Therm Biol. 1983;8(1-2):143–147. [Google Scholar]
- Nagashima K, Matsue K, Konishi M, Iidaka C, Miyazaki K, Ishida N. et al. The involvement of Cry1 and Cry2 genes in the regulation of the circadian body temperature rhythm in mice. Am J Physiol Regul Integr Comp Physiol. 2005;288(1):R329–R335. doi: 10.1152/ajpregu.00395.2004. [DOI] [PubMed] [Google Scholar]
- Um J. et al. AMPK regulates circadian rhythms in a tissue- and isoform-specific manner. PLoS One. 2011;6(3):e18450. doi: 10.1371/journal.pone.0018450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, Andrews JL. et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci USA. 2007;104(9):3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JJ, Andrews JL, McDearmon EL, Campbell KS, Barber BK, Miller BH. et al. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics. 2007;31(1):86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P, Nixon SJ, Parton RG, Muscat GE. RORalpha regulates the expression of genes involved in lipid homeostasis in skeletal muscle cells: Caveolin-3 and CPT-1 are direct targets of ROR. J Biol Chem. 2004;279(35):36828–36840. doi: 10.1074/jbc.M404927200. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan SN, Lau P, Burke LJ, Muscat GE. Rev-erbbeta regulates the expression of genes involved in lipid absorption in skeletal muscle cells: Evidence for cross-talk between orphan nuclear receptors and myokines. J Biol Chem. 2005;280(10):8651–8659. doi: 10.1074/jbc.M413949200. [DOI] [PubMed] [Google Scholar]
- Kamei Y, Ohizumi H, Fujitani Y, Nemoto T, Tanaka T, Takahashi N. et al. PPARgamma coactivator 1beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc Natl Acad Sci USA. 2003;100(21):12378–12383. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): Transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24(1):78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- Andrews JL, Zhang X, McCarthy JJ, McDearmon EL, Hornberger TA, Russell B. et al. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc Natl Acad Sci USA. 2010;107(44):19090–19095. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauermann R, Schmidt WM, Krebs M, Brunner M, Muller M. Ramipril modulates circadian gene expression in skeletal muscle. Pharmacogenet Genomics. 2011;21(11):751–759. doi: 10.1097/FPC.0b013e32834a8621. [DOI] [PubMed] [Google Scholar]
- Muoio DM, Newgard CB. Mechanisms of disease: Molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9(3):193–205. doi: 10.1038/nrm2327. [DOI] [PubMed] [Google Scholar]
- Polonsky KS, Given BD, Van Cauter E. Twenty-four-hour profiles and pulsatile patterns of insulin secretion in normal and obese subjects. J Clin Invest. 1988;81(2):442–448. doi: 10.1172/JCI113339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polonsky KS, Given BD, Hirsch LJ, Tillil H, Shapiro ET, Beebe C. et al. Abnormal patterns of insulin secretion in non-insulin-dependent diabetes mellitus. N Engl J Med. 1988;318(19):1231–1239. doi: 10.1056/NEJM198805123181903. [DOI] [PubMed] [Google Scholar]
- Peschke E, Peschke D. Evidence for a circadian rhythm of insulin release from perifused rat pancreatic islets. Diabetologia. 1998;41(9):1085–1092. doi: 10.1007/s001250051034. [DOI] [PubMed] [Google Scholar]
- Muhlbauer E, Wolgast S, Finckh U, Peschke D, Peschke E. Indication of circadian oscillations in the rat pancreas. FEBS Lett. 2004;564(1-2):91–96. doi: 10.1016/S0014-5793(04)00322-9. [DOI] [PubMed] [Google Scholar]
- Allaman-Pillet N, Roduit R, Oberson A, Abdelli S, Ruiz J, Beckmann JS. et al. Circadian regulation of islet genes involved in insulin production and secretion. Mol Cell Endocrinol. 2004;226(1-2):59–66. doi: 10.1016/j.mce.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH. et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466(7306):627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadacca LA, Lamia KA, deLemos AS, Blum B, Weitz CJ. An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia. 2011;54(1):120–124. doi: 10.1007/s00125-010-1920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira E, Marroqui L, Batista TM, Caballero-Garrido E, Carneiro EM, Boschero AC. et al. The clock gene rev-erbalpha regulates pancreatic beta-cell function: Modulation by leptin and high-fat diet. Endocrinology. 2012;153(2):592–601. doi: 10.1210/en.2011-1595. [DOI] [PubMed] [Google Scholar]
- Fonken LK, Nelson RJ. Illuminating the deleterious effects of light at night. F1000 Med Rep. 2011;3:18. doi: 10.3410/M3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack RL. The pathophysiology of jet lag. Travel Med Infect Dis. 2009;7(2):102–110. doi: 10.1016/j.tmaid.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci USA. 2009;106(11):4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XS, Armstrong ME, Cairns BJ, Key TJ, Travis RC. Shift work and chronic disease: The epidemiological evidence. Occup Med (Lond) 2011;61(2):78–89. doi: 10.1093/occmed/kqr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G. Calorie restriction increases life span: A molecular mechanism. Nutr Rev. 2006;64(2 Pt 1):89–92. doi: 10.1301/nr.2006.feb.89-92. [DOI] [PubMed] [Google Scholar]
- Challet E. Interactions between light, mealtime and calorie restriction to control daily timing in mammals. J Comp Physiol B. 2010;180(5):631–644. doi: 10.1007/s00360-010-0451-4. [DOI] [PubMed] [Google Scholar]
- Froy O, Miskin R. Effect of feeding regimens on circadian rhythms: Implications for aging and longevity. Aging (Albany NY) 2010;2(1):7–27. doi: 10.18632/aging.100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praet SF, van Loon LJ. Exercise therapy in type 2 diabetes. Acta Diabetol. 2009;46(4):263–278. doi: 10.1007/s00592-009-0129-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G, Esser KA. Scheduled exercise phase shifts the circadian clock in skeletal muscle. Med Sci Sports Exerc. 2012;44(9):1663–1670. doi: 10.1249/MSS.0b013e318255cf4c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermida RC, Ayala DE, Mojon A, Fernandez JR. Influence of time of day of blood pressure-lowering treatment on cardiovascular risk in hypertensive patients with type 2 diabetes. Diabetes Care. 2011;34(6):1270–1276. doi: 10.2337/dc11-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggs JE, Hogenesch JB. Genomics and systems approaches in the mammalian circadian clock. Curr Opin Genet Dev. 2010;20(6):581–587. doi: 10.1016/j.gde.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Grant GR, Paquin C, Qian J, Nitabach MN. Deep sequencing the circadian and diurnal transcriptome of drosophila brain. Genome Res. 2012;22(7):1266–1281. doi: 10.1101/gr.128876.111. [DOI] [PMC free article] [PubMed] [Google Scholar]