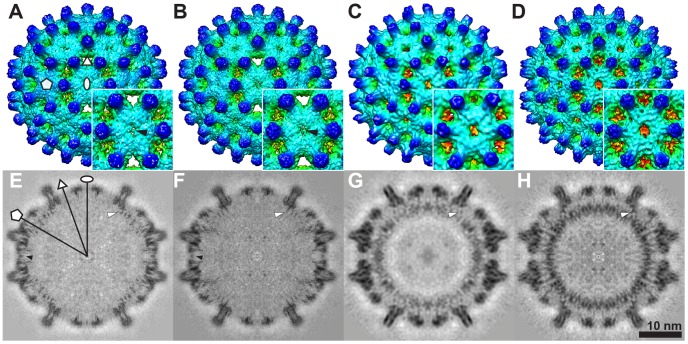
Figure 2. Cryo-EM 3D reconstructions of empty and pgRNA-filled Cp183 capsids.
Surface shaded exterior maps of T = 4 (A) Cp183e-SSS, (B) Cp183e-EEE, (C) Cp183RNA-SSS, (D) Cp183RNA-EEE and their related central sections (E–H). Insets show enlarged views of the twofold (i.e. quasi-sixfold) vertex. All four maps have a similar external appearance with 120 spikes decorating a fenestrated capsid surface; the outer layer extends from a radius of 125 to 170 Å. In (A) Cp183e-SSS and (B) Cp183e-EEE, a thin layer of electron density partially occludes the central opening pore at the twofold axis (A, B, E, F, black arrows). This density is unique to the empty capsids. The pgRNA-filled capsids (C, D, G, H), on the other hand, lack the density across the twofold pore but display a substantial internal layer of density at the radii between 100–120 Å. In central sections (G, H), this density, corresponding to the co-assembled pgRNA, is clearly inhomogeneous indicating that the pgRNA has adopted a preferred conformation or constellation of conformations evident even though it has been icosahedrally averaged in these reconstructions. White arrows indicate the CTD tails tethered from the capsid inner surface. Oval, triangle, and pentagon indicate locations of twofold, threefold and fivefold axes, respectively.