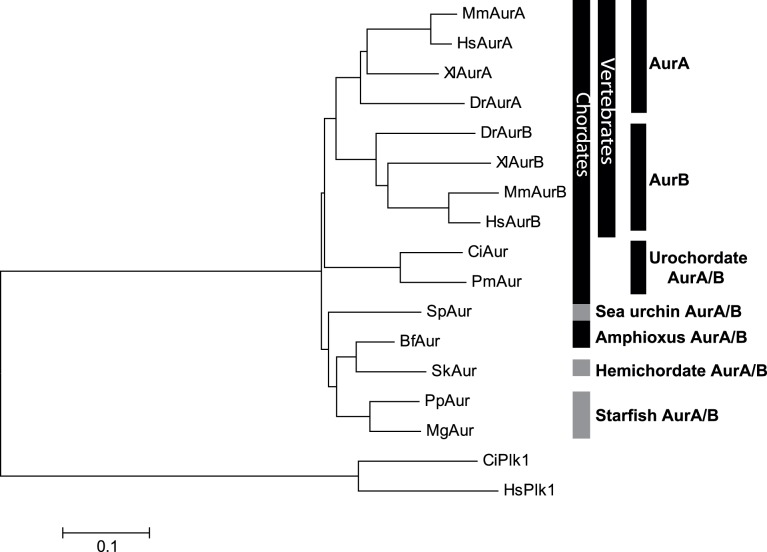
Figure 6. Phylogenetic tree of deuterostome Aurora-A, Aurora-B and Aurora-AB kinases rooted by Plk1 kinases.
Phylogenetic tree constructed using CLUSTALW in BioEdit to align the catalytic domains of deuterostome Auroras. The tree was produced using MEGA employing the neighbor joining method. Species represented :HsAur A and B (human), MsAur A and B (mouse), XlAur A and B (Xenopus laevis), DrAur A and B (Danio rerio), CiAur (Ciona intestinalis, ascidian), PmAur (Phallusia mammillata, ascidian), SpAur (Strongylocentrotus purpuratus, sea urchin), SkAur (Saccoglossus kowalevskii, hemichordate), BfAur (Branchiostoma floridae, cephalochordate), MgAur (Marthasterias glacialis, starfish), and PpAur (Patiria pectinifera, starfish). Human and Ciona Polo like kinase 1 were used as the outgroup.