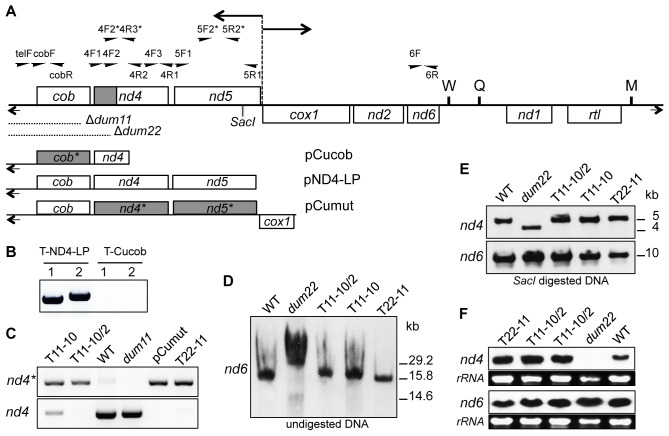
Figure 1. Molecular characterization of the transformants.
(A) Schematic map of the Chlamydomonas reinhardtii mitochondrial genome. Boxes represent protein-coding genes: (cob) apocytochrome b of complex III; (nd1, 2, 4, 5 and 6) subunits of complex I; (cox1) subunit 1 of complex IV; (rtl) reverse transcriptase-like protein. W, Q, and M represent tRNAs for Trp, Glu, and Met, respectively. The bidirectional origin of transcription between nd5 and cox1 genes is represented by a dashed vertical line and two horizontal arrows. Terminal inverted repeats are shown by short arrows and SacI digestion site at position 5.5 kb (GenBank u03843 numbering) is indicated. Region where modifications on the nd4 gene were found on T11-10, T11-10/2 and T22-11 transformants is indicated in grey. Position and name of primers are indicated above the map. Primers with a star are specific for the modified gene version (for primer sequence see Table S1). Positions of the dum11 and dum22 deletions are shown. Mitochondrial DNA fragments contained in pND4-LP, pCucob and pCumut are schematized. Grey boxes represent the modified genes where GGC/GGT codons were changed in GGG codons. (B) Detection of the cob gene in transformants obtained after biolistic transformation with pND4-LP (T-ND4-LP) and pCucob (T-cucob) constructs. PCR analyses were performed with cobF/cobR (1) and telF/cobR (2) pair primers. (C) Detection of the mutated and the wild-type nd4 genes on T11-10, T11-10/2 and T22-11 transformants. PCR analyses were performed with 4F2*/4R2 and 4F2/4R2 pair primers for the modified nd4 gene (nd4*) and for wild-type nd4 gene (nd4) respectively. (D–E) Reconstitution of complete mitochondrial genome in T11-10, T11-10/2 and T22-11 transformants. Southern blot analyses were performed (D) on total DNA with the nd6 PCR probe and (E) on SacI digested DNA with nd4 and nd6 PCR probes. (F) Transcript levels of nd4 and nd6 genes in T11-10, T11-10/2 and T22-11 transformants. Northern blot analyses were performed on total RNA with nd4 and nd6 PCR probes. Loadings of rRNAs are shown.