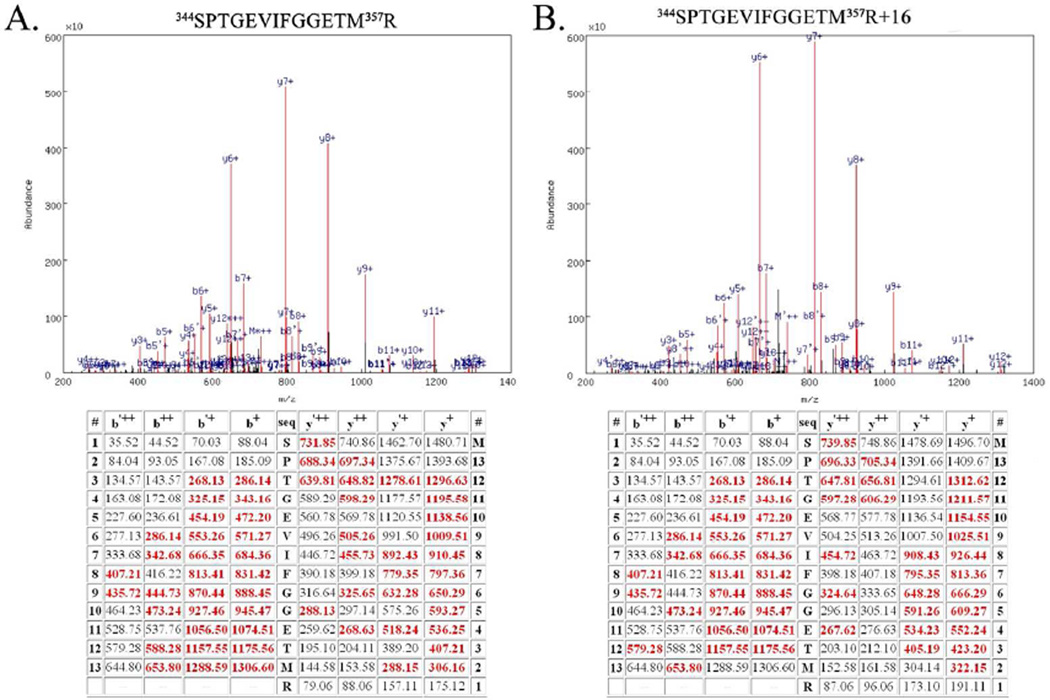
Figure 1.
Example Mass Spectrometry Data from the Unmodified Peptide 344SPTGEVIFGGETM357R and the Oxidatively Modified Peptide 344SPTGEVIFGGETM357R+16. A. Top, spectrum of the CID dissociation of the unmodified peptide 344SPTGEVIFGGETM357R. Various identified ions are labeled. Bottom, table of all predicted masses for the y- and b- ions generated from this peptide sequence. Ions identified in the CID spectrum (above) are shown in red. The b’++, b’+ y’++ and y’+ ions are generated by the neutral loss of water. B. Top, spectrum of the CID dissociation of the modified peptide344SPTGEVIFGGETM357R+16. Various identified ions are labeled. Bottom, table of all predicted masses for the y- and b- ions generated from this peptide sequence. Ions identified in the CID spectrum are shown in red. The b’++, b’+ y’++ and y’+ ions are generated by the neutral loss of water. Note the additional mass of 16 amu (in comparison with Fig. 1A) arising from modification of 357R. This modification is evident in all identified y-ions species but is absent in all identified b-ions. Both of these spectra are from Biological Replicate #1.