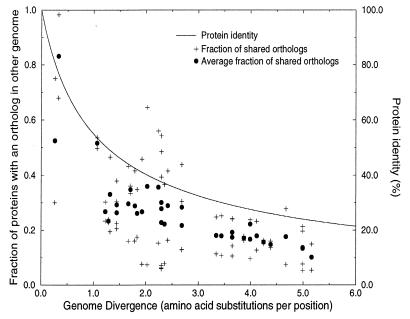
Figure 2.
The relationship between genome similarity, measured as the fraction of shared orthologs, and time, measured as the number of amino acid substitutions per protein per position in a set of 34 orthologs. + shows the fraction of sequences in a genome A that has an ortholog in another genome B, and vice versa. This measure is asymmetric, a relatively small genome like H. influenzae is more similar to a large one like E. coli than E. coli is similar to H. influenzae. • shows the average of the two asymmetric similarities. Here we use a minimal definition of orthology: sequences that between two genomes have the highest, significant (E < 0.01) level of pairwise identity, that covers at least 60% of one of the proteins are regarded as orthologs. Sequences were compared with the Smith–Waterman algorithm (47), using a parallel Bioccellerator computer. The relationship between sequence identity and the number of amino acid substitutions per position as calculated with Grishin’s equation (25) is given for comparison. If one assumes that the divergence time between the Archaea and Bacteria is 3.5 billion years (23), the unit of one amino acid substitution corresponds to about 875 million years. In this estimate of divergence time the Mycoplasmas and H. pylori are not included, because they have a relatively high rate of evolution. The highest six divergence times correspond to the comparisons of the Mycoplasmas and H. pylori with the Archaea. As is clear from the figure, the fraction of shared orthologs between genomes decreases more rapidly in evolution than does the protein identity. Note that the base level of shared orthologs at which the figure saturates consists only partly of a set of sequences that are shared by all the genomes compared. For example, there are 15 orthologous pairs shared between M. genitalium and M. thermoautotrophicum of which none of the genes has a homolog at the E < 0.01 level in M. jannaschii. Of this set, the ones with the highest level of protein identity are: DnaK and DnaJ (MG305 and MG019), heat shock proteins with 51% and 50% identity, respectively to their M. thermoautotrophicum ortholog, deoxyribose-phosphate aldolase (MG050) with 40% identity, a pyrophosphatase (MG351) with 40.5% identity, and a transcriptional regulator (MG448) with 45% identity. Genes that are shared by M. genitalium and M. jannaschii but that are absent in M. thermoautotrophicum, include proteins from the glycolysis like pyruvate kinase (MG216) with 29.1% identity and glucose-6-phosphate isomerase (MG111) with 27% protein identity.