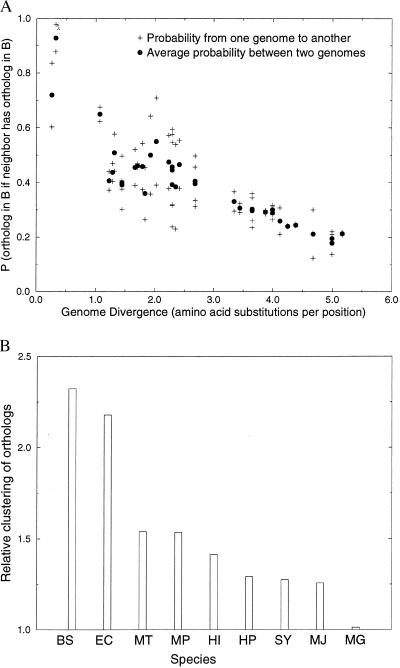
Figure 5.
(A) The probability that a gene in genome A has an ortholog in another genome B if a neighboring gene in A has an ortholog in genome B. The probabilities clearly increase, as compared with the average probability of having an ortholog in another genome (compare Fig. 2). (B) The relative degree of clustering of genes in one genome (A) that have an ortholog in another genome (B). The analysis includes only genes that are clustered (“neighbors”) in genome A, but not in B (and vice versa). Shown is the ratio of the number of genes in A that have an ortholog in B and have at least one neighboring gene that also has an ortholog in B, divided by the expected number. The expected number of genes that are neighbors in a genome, given a random distribution, is calculated as follows: Given X genes that are randomly distributed over a genome with Y loci, the probability that a gene from X has no neighboring genes from X (it lies isolated) is the probability that it has no left-neighbor from X nor a right-neighbor from X: P0 = [(Y − X)/(Y − 1)]* [(Y − X − 1)/(Y − 2)]. The expected number of genes from X with at least one neighbor from X: P1,2 = 1 − P0. The fraction of genes in genome A with at least one neighbor that also has an ortholog in genome B is thus divided by P1,2 to get to the relative clustering of the genes in genome A. The relative clustering is averaged over the genome comparisons of one genome versus the eight other genomes. The names of the species have been abbreviated to the first letters of their genus and species name. All genomes, except M. genitalium show a more than expected clustering of genes. Given its small size, M. genitalium has relatively little room to cluster the genes that have an ortholog in another genome above the expected level of clustering: i.e., most of the genes that have an ortholog in another genome are expected to be neighbors in M. genitalium. The correlation with genome size is not perfect however. For example, Synechocystis, which has a relatively large genome, shows relatively little genome organization.