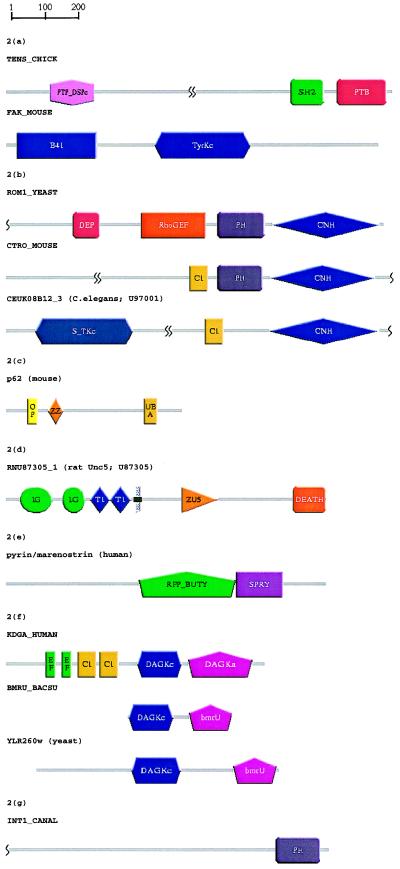
Figure 2.
Schematic representations, produced using SMART, of the domain architectures of proteins discussed in the text. See Table 1 for the identified domains; gray lines (no SMART match) might contain other known domains not included in SMART. Putative homologues were identified during SWise (16) searches and/or psi-blast (1) searches (E < 0.01). (a) Domain recognition: A novel PTB domain was identified in tensin, resulting in completion of its modular architecture assignment. A psi-blast search with a previously predicted PTB domain in C. elegans F56D2.1 (53) yields the tensin PTB after four passes. Prediction of molecular function via domain hit: Identification of a domain homologous to band 4.1 protein in focal adhesion kinase (FAK) isoforms. FAKs are predicted to bind cytoplasmic portions of integrins in a similar manner to that of talin, another band 4.1 domain-containing protein. A psi-blast search with a band 4.1-like domain (41 HUMAN, residues 206–401) revealed band 4.1-like domains in human, bovine, and Xenopus FAK isoforms by pass 3. (b) Detection of new domains because of search space reduction: Putative DEP domains in ROM1 and ROM2 were identified by using SWise (16) and HMMer (14), but could not be detected by using psi-blast. Analysis of the regions surrounding identified domains revealed the presence of a novel domain in the C-terminal regions of ROM1 and ROM2 that occurs also in several Ste20-like protein kinases, and mouse citron (CNH, citron homology). A gapped blast search of the region of citron C-terminal to its PH domain (CTRO MOUSE, residues 1134–1457) reveals significant similarity with yeast ROM2 (E = 1 × 10−5). (c) Functional predictions for an entire domain family: A region of p62 known to bind ubiquitin (40), and its homologous sequence in the Drosophila protein ref(2)P, scored as the highest putative true negatives in a SWise search. We predict ubiquitin-binding functions for UBA domains. psi-blast searches were unable to corroborate this prediction. (d) Prediction of cellular functions: Although not indicated in the primary sources (43, 44), a DEATH domain was found in rcm and other UNC5 homologues, in agreement with a previous claim (41). At the molecular level, this domain in UNC5 is predicted to form a heterotypic dimer with an homologous domain in UNC44 implying a cellular role in axon guidance. A gapped blast search with the known DEATH domain of death-associated protein kinase (DAPK HUMAN, residues 1304–1396) predicts a DEATH domain in rat UNC5H1 with E = 9 × 10−3). (e) Signaling domains in “disease genes”: Pyrin or marenostrin, a protein that is mutated in patients with Mediterranean fever and is similar to butyrophilin, contains a SPRY domain. psi-blast with the SPRY domain of human DDX1 (EMBL:X70649, residues 124–240) yields a butyrophilin homologue by pass 5 and pyrin/marenostrin (residues 663–759) by pass 7. (f) Homologues of domains involved in eukaryotic signaling may not be eukaryotic-specific: DAG kinases have been found previously in mammals, invertebrates, plants, and slime mold. However, it is apparent that DAG kinase homologues of unknown function are present in yeasts and in eubacteria (see Fig. 3). A gapped blast search with Bacillus subtilis bmrU (BMRU BACSU) yields significant similarities with Arabidopsis thaliana DAG kinase (KDG1 ARATH; E = 4 × 10−4) and a Schizosaccharomyces pombe ORF (SPAC4A8.07c; E = 1 × 10−7). (g) Identification of potential misclassifications: A PH domain and the lack of an obvious transmembrane sequence indicates a cytoplasmic and signaling role for a protein (INT1 CANAL) previously thought to be a yeast integrin. A psi-blast search with the N-terminal PH domain of pleckstrin yielded INT1 CANAL in pass 3.