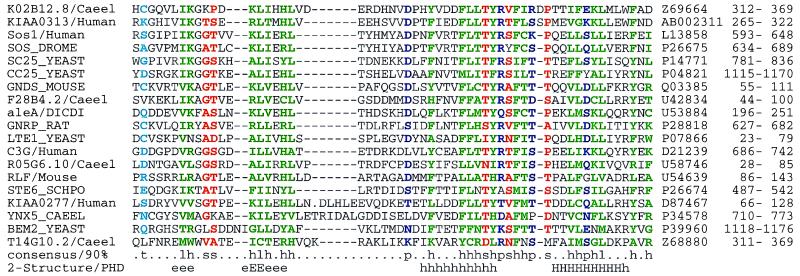
Figure 3.
Multiple alignments of selected RasGEFN domains. A conserved region was found in the N-terminal regions of several proteins with RasGEF (Cdc25-like) domains (37). Surprisingly, this N-terminal domain may be present in the sequence either close to, of far from, the RasGEF domain. A psi-blast search using a region (residues 898–946) of C. albicans Cdc25 (CC25 CANAL) and E < 0.01, identified each of the sequences in Fig. 3 within nine passes before convergence. Predicted (54) secondary structure and 90% consensus sequences are shown beneath the alignments; SwissProt/PIR/EMBL accession codes and residue limits are given after the alignments. Residues are colored according to the consensus sequence [green: hydrophobic (h), ACFGHIKLMRTVWY; blue: polar (p), CDEHKNQRST; red: small (s), ACDGNPSTV; red: tiny (u), AGS; cyan: turn-like (t), ACDEGHKNQRST; green: aliphatic (l), ILV; and, magenta: alcohol (o), ST). The SwissProt sequence KMHC DICDI has been altered to account for probable frameshifts.