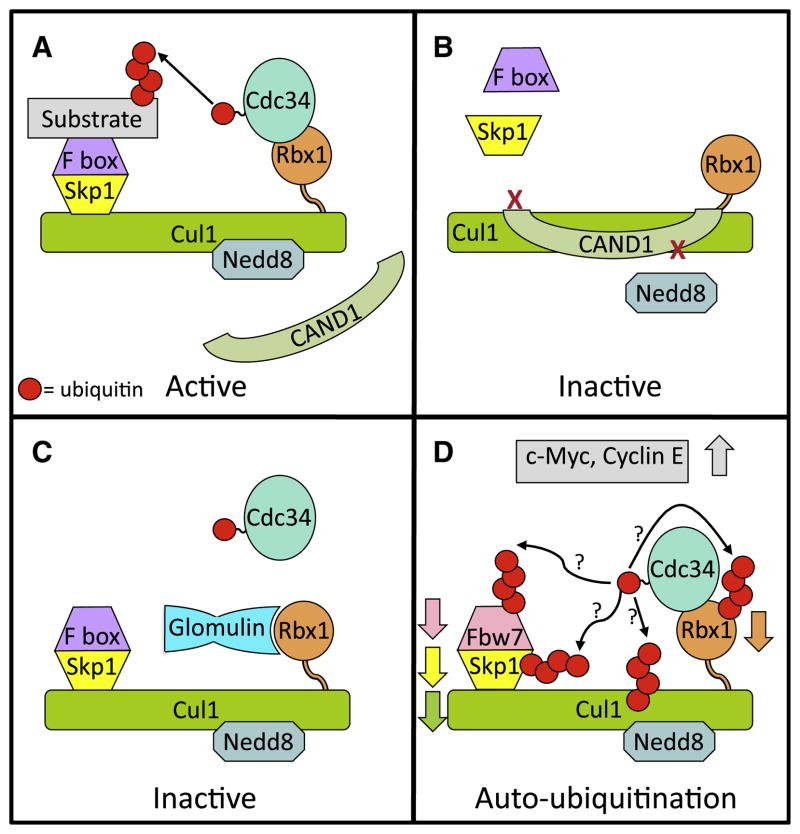
Figure 1. Regulation of CRL1.
(A) Functional CRL1. Cullin RING ligase 1 (CRL1) family members are comprised of Cul1, the RING finger Rbx1, which binds the active E2 (Cdc34~Ub), the adaptor protein Skp1, and one of many substrate-binding F box proteins. Activation of CRL1 requires modification of the cullin core with the ubiquitin-like protein Nedd8, which initiates assembly of the Skp1 and F box proteins on the CRL complex and prevents association of the CRL inhibitor CAND1.
(B) CRL1 inhibition through complex disassociation. Binding of the CRL inhibitor CAND1 to Cul1-Rbx1 inhibits neddylation and prevents assembly of the Skp1 F box complex.
(C) Inhibition of CRL1 activity by glomulin. A recently discovered mechanism for CRL inhibition is by association of the protein glomulin with Rbx1, preventing the RING finger of Rbx1 from binding to Cdc34~Ub.
(D) Speculative model of the effects of loss of glomulin. Absence of glomulin results in decreased levels of the F box protein Fbw7, Rbx1, and Cul1 through ubiquitination and proteasomal degradation leading to accumulation of the CRL1Fbw7 substrates c-Myc and Cyclin E. This is likely to occur through enhanced autoubiquitination of CRL1 components.