Abstract
Loss of heterozygosity (LOH) studies indicate that genetic alterations of chromosome 9p occur in numerous tumor types, suggesting the presence of tumor suppressor genes (TSGs) on chromosome 9p critical in carcinogenesis. Our previous LOH analyses in primary lung tumors led us to propose that chromosome 9p harbors other TSGs important in lung tumorigenesis. In this study, 30 non-small-cell lung cancer and 12 small-cell lung cancer cell lines were screened with 55 markers to identify new regions of homozygous deletion (HD) on chromosome 9p. Three novel noncontiguous homozygously deleted regions were detected and ranged in size from 840 kb to 7.4 Mb. One gene identified in the deletion at D9S126, TUSC1 (tumor suppressor candidate 1), is an intronless gene. Multiplex polymerase chain reaction and Southern blot confirmed the HD of TUSC1. Northern blot analysis of TUSC1 demonstrated two transcripts of approximately 2 and 1.5 kb that are likely generated by alternative polyadenylation signals. Both transcripts are expressed in several human tissues and share an open-reading frame encoding a peptide of 209 amino acids. Analysing cell line cDNAs by reverse transcriptase (RT)–PCR demonstrated downregulation of TUSC1 in cell lines with or without HDs, suggesting that TUSC1 may play a role in lung tumorigenesis.
Keywords: homozygous deletion, chromosome 9, lung cancers, TUSC1, expression, tumor suppressor gene
Introduction
Lung cancer is the leading cause of cancer-related mortality in the United States as well as a world-wide public health problem (Oh et al., 2002). Chromosome 9p has been implicated in chromosomal inversions, translocations, loss of heterozygosity (LOH), and homozygous deletion (HD) in a variety of malignant cell lines including melanoma, non-small-cell lung carcinoma (NSCLC), breast cancer, leukemia, and hepatocellular carcinoma. These data suggest chromosome 9p contains a tumor suppressor locus (loci) that may be involved in the genesis of several tumor types (Cheng et al., 1993; Coleman et al., 1994; Holland et al., 1994; Kamb et al., 1994; An et al., 1996; Mead et al., 1997; Takeuchi et al., 1997; Wiest et al., 1997; Sheu et al., 1999; Pollock et al., 2001). Two candidate tumor suppressor loci have been identified in the chromosome 9p21 region, one is p16/CDKN2A (INK4A or MTS1) that encodes the p16 and p14ARF proteins and the other one is p15/CDKN2B (INK4B or MTS2) that encodes the p15 protein (Serrano et al., 1993; Kamb et al., 1994; Quelle et al., 1995). Since p16/CDKN2A is frequently inactivated genetically or epigenetically in cancer cells, the p16/CDKN2A locus is suspected to be a major tumor suppressor gene (TSG) (Cairns et al., 1995; Merlo et al., 1995; Xiao et al., 1995; Castellano et al., 1997; Hamada et al., 1998; Hamada et al., 2000).
Previous studies with squamous cell carcinoma of the lung in our laboratory identified a region of HD at the microsatellite marker D9S126, which is distinct from the p16/CDKN2A locus and lies approximately 4 Mb proximal to p16/CDKN2A (Wiest et al., 1997). These findings suggested other regions of HD harboring additional TSG(s) important in lung tumorigenesis may be present on chromosome 9p. Other reports support this conclusion (Hamada et al., 2000).
In the present study, we have screened 30 NSCLC and 12 small-cell lung carcinoma (SCLC) cell lines with 55 sequence-tagged sites (STS) or microsatellite markers. The markers, mapped both distal and proximal to the p16/CDKN2A locus, were used to search for unknown HDs and to define additional tumor suppressor loci on chromosome 9p.
Results
HD mapping of chromosome 9p
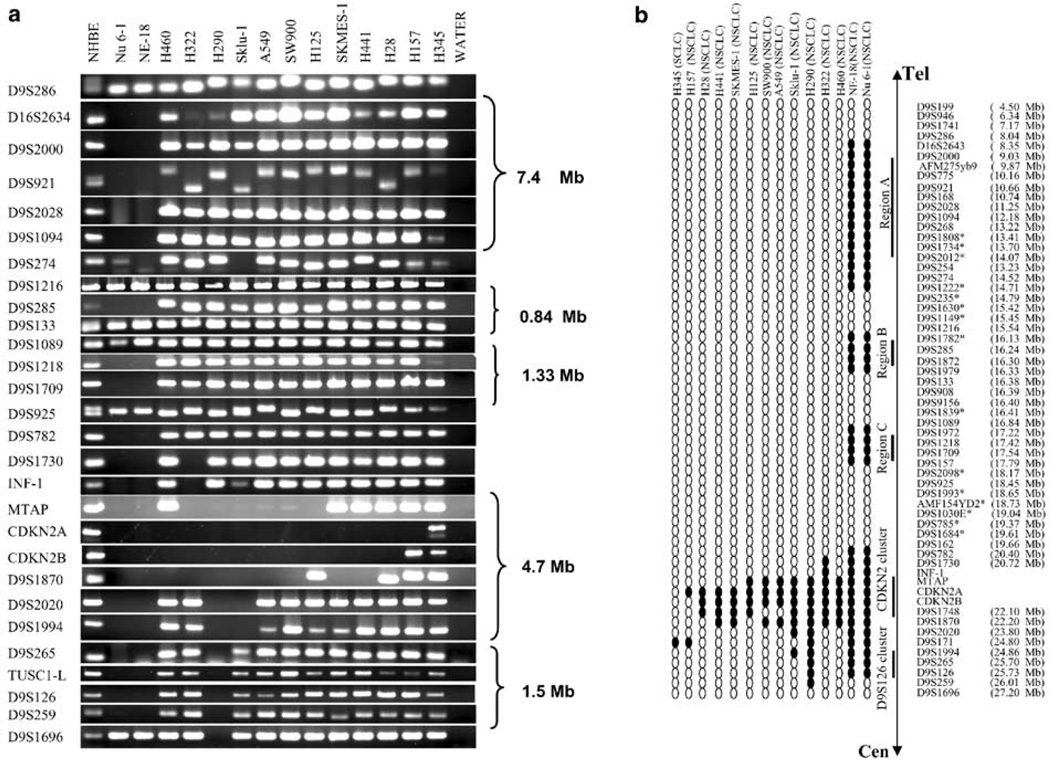
A novel homozygously deleted region at marker D9S126 in squamous cell carcinoma of the lung indicated the possibility of additional homozygously deleted loci on short arm of chromosome 9 (Wiest et al., 1997). To identify potential lung cancer TSG(s) in these regions, we expanded our search for novel deletion region(s) within the DNA of 42 lung cancer cell lines using polymerase chain reaction (PCR) with primers for four genes located in or close to the p16/CDKN2 locus and 55 STSs or microsatellite markers (Figure 1a and b). Deletions of the interferon-omega 1 (INF-1) and methylthioadenosine phosphorylase (MTAP) genes together with the deletion of p16/CDKN2A occurred in most cases (Figure 1a and b). Two frequently deleted regions, one at the p16/CDKN2A locus in 13 NSCLC cell lines and the other one at D9S126, were identified. However, this HD was contiguous with the deletion of p16/CDKN2A in three NSCLC cell lines, NE-18, Nu 6-1, and H290 (Figure 1a and b). In addition, three noncontiguous HDs distal to p16/CDKN2A, designated regions A–C (Figure 1b), were detected in the two NSCLC variant cell lines, NE-18 and Nu 6-1. Cell lines H345 (SCLC) and H157 (NSCLC) showed minimal interstitial deletions at D9S171, and Sklu-1 showed interstitial deletion of D9S1994 (Figure 1a and b). Moreover, these deletions were not contiguous with the deletion of CDKN2 locus. Representative agarose gels of HDs are shown (Figure 1a). Based on the location of the STSs or microsatellite markers, the approximate sizes of three deletion regions are 7.4 Mb for region A defined by D9S286 and D9S235, 840 kb for region B defined by D9S1216 and D9S133, and 13 Mb for region C defined by D9S1089 and D9S2098 (Figure 1b).
Figure 1.
HDs of chromosome 9p in lung cancer cell lines. (a) PCR analysis of STS or microsatellite markers in lung cancer cell lines. All markers are arranged from telomeric (D9S199) to centromeric (D9S1696). INF-1, MTAP, exon 2 of CDKN2A and CDKN2B loci are shown together with the DNA makers. The approximate sizes of the deletions are based on the database (http://www.ensembl.org/). (b) HD map of the 9p21 region. Results of PCR analysis for each STS or microsatellite is shown by filled oval representing HD and open oval as retention. Two HDs clustered at p16/CDKN2A and D9S126 and three novel regions A–C were indicated. Approximate sequence positions of DNA markers in the database (http://www.ensembl.org/) were shown. NSCLC: non-small-cell lung carcinoma, SCLC: small-cell lung carcinoma. *DNA markers were only performed on samples of NE-18, Nu 6-1, and NHBE
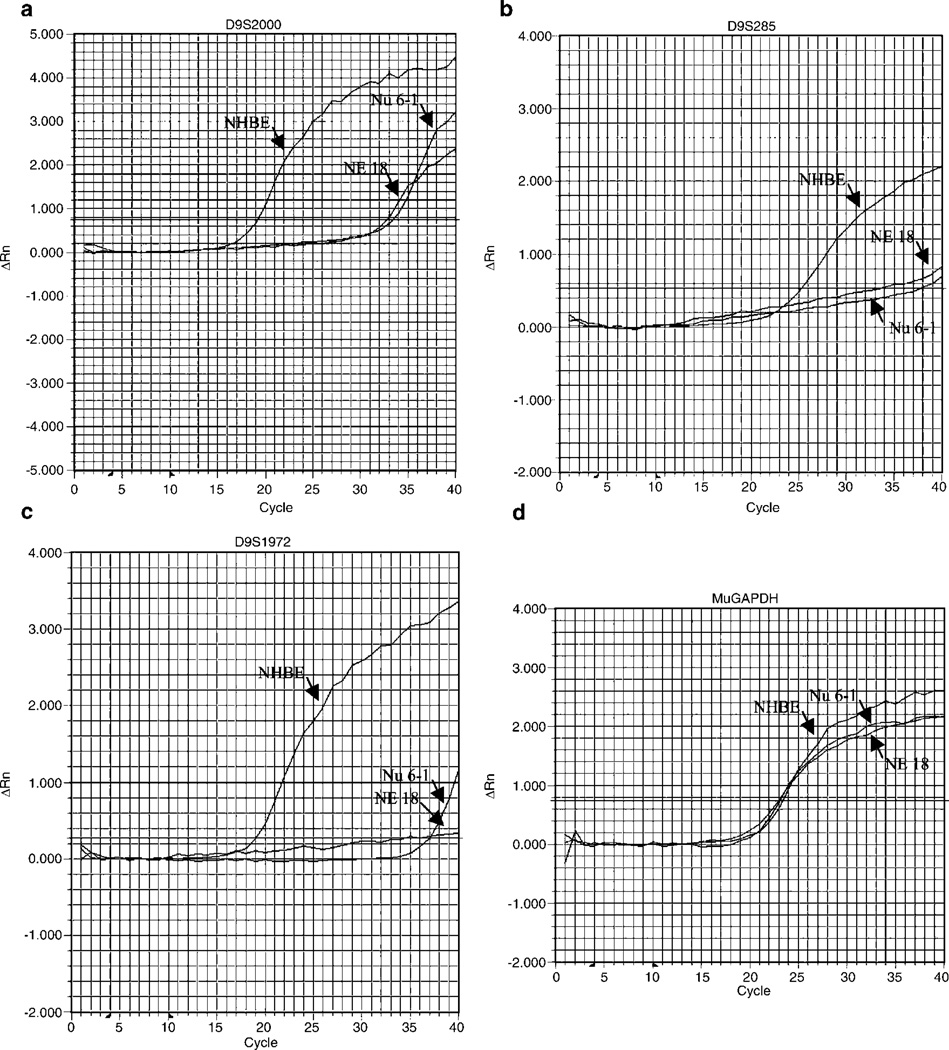
We performed quantitative real-time polymerase chain reaction (PCR) analysis with nine markers within or close to the novel deleted regions to further confirm the HDs detected in both NE-18 and Nu 6-1. The relative copy numbers for deleted or retained DNA markers in tumor and control DNA were listed in Table 1, and the representative amplification plots clearly confirm our PCR results (Figure 2).
Table 1.
Relative copy number of DNA markers in NE-18 and Nu 6-1 compared with an NHBE
| Microsatellite | NHBEa | NE-18a | Nu 6-1a | PCR status |
|---|---|---|---|---|
| D9S286 | 1 | 0.1 | 0.2 | Retention |
| D9S2000 | 1 | 0.00009 | 0.0001 | HD |
| D9S1094 | 1 | 0.015596 | 0.0062 | HD |
| D9S1216 | 1 | 1.2435 | 1.2812 | Retention |
| D9S285 | 1 | 0.0002 | 0.0002 | HD |
| D9S133 | 1 | 0.7206 | 0.6314 | Retention |
| D9S1972 | 1 | 0.000004 | 0.0004 | HD |
| D9S1218 | 1 | 0.0001 | 0.0001 | HD |
| D9S1993 | 1 | 0.482 | 0.4381 | Retention |
Values are determined by real-time PCR and calculated as described in ‘Materials and methods’ using the comparative CT methods
Figure 2.
Quantative real-time PCR amplification in cell lines NE-18, NU 6-1, and NHBE. Amplification plots of representative real-time PCR analyses for the DNA markers and MuGAPDH in tumor DNA (NE-18 and Nu 6-1) compared with control DNA (NHBE). D9S2000 is within deleted region A, D9S285 is within deleted region B and D9S1218 is within deleted region C
Sequence analyses of human TUSC1 (tumor suppressor candidate 1) and mouse Tusc1
Systematic characterization of genes located in a HD region in cancer cell lines was a potential approach for identifying TSGs important in tumorigenesis. Since the frequency of HD at the D9S126 locus is higher than the other three regions defined in this study and was also identified in our previous study of primary tumors, we initiated a molecular analysis identifying potential TSGs from this region (Figure 1b). A 1 Mb BAC clone contig (data not shown) of chromosome 9p21 including the distal marker D9S265 and proximal marker D9S259 containing the marker D9S126 was derived from the database. These clones were used to identify genes located in this HD region (Wiest et al., 1997). We first analysed the available genome sequences of this region and found three predicted exon sequences (data not shown) that were selected for further investigation. Database searching indicated that Loc169556 (NCBI) was highly similar to several ESTs, for example, I.M.A.G.E. 4867018 and 5169320 and C75430 from Stratagene (La Jolla, CA, USA). Sequence analysis indicated that one EST clone, I.M.A.G.E. 5169320, was 1473 bp, contained a potential initiation codon, and had a consensus AATAA polyadenylation signal 24 bp upstream of the poly-A+ tail in the 3′-untranslated region (UTR). Further database analysis indicated that the sequences of the majority of related EST clones were identical to the sequence of I.M.A.G.E. 5169320 and shared the same polyadenylation signals (data not shown). However, there was an EST clone (I.M.A.G.E. 815591) whose sequence was identical to I.M.A.G.E. 5169320 except for an additional 574 bps located downstream of the first polyadenylation signal (Figure 3a). A single fragment was amplified by PCR with TUSC1-S1 and TUSC1-L1 from the cDNA generated from the normal human bronchial/tracheal epithelial cell culture (NHBE) cell line. Sequence comparison of PCR products indicated that the amplicons were identical to the corresponding clones, I.M.A.G.E. 5169320 and I.M.A.G.E. 815591. These results suggest both clones belong to the same gene or gene family, which we refer to as TUSC1 from the HUGO Gene Nomenclature Committee (HGNC) and the TUSC1 locus codes for different transcripts generated by alternative polyadenylation signals. The composite cDNA sequence of I.M.A.G.E. 5169320 and I.M.A.G.E. 815591 as TUSC1-L (TUSC1 Long) (Figure 3) and was deposited in GenBank (Accession no.: AY168647), while the sequence of I.M.A.G.E. 5169320 was referred to as TUSC1-S (TUSC1 Short). It should also be noted that TUSC1-L and TUSC1-S share the same open-reading frame (ORF) (Figure 3a) of 627 nucleotides, encoding a 209 amino-acid polypeptide (Figure 3a). No conserved protein domain(s) were found in the protein sequence. Further searching of the human genome indicated that the entire cDNA sequence was found in a single BAC clone, RP11-536F2, which neighbors the BAC clone RP11-33G16 containing D9S126. In addition, the cDNA sequence was contiguous with the genomic sequence, suggesting that TUSC1 was intronless and located in the HD at D9S126.
Figure 3.
(a) Nucleotide sequence and deduced amino-acid sequence of human TUSC1. Nucleotides are numbered with respect to the first nucleotide of the translation initiation codon. Amino acids of the ORF are represented by the one-letter code below the cDNA nucleotide sequence. The translation stop codon is indicated by an asterisk (*) and putative polyadenylation signals in the 3′-UTR are underlined. (b) Comparison of amino-acid sequences of human TUSC1 and mouse Tusc1
We performed database searches for the mouse homologue using the sequence of TUSC1-L to substantiate the existence of the human gene. Two mouse I.M.A.G.E. clones, 3495226 and 5033231, were identified and sequenced. Sequence analysis showed that I.M.A.G.E. clone 3495226 contains 1157 bps with a putative ORF and a consensus AATAA polyadenylation signal 12 bps upstream of the poly-A+ tail in the 3-UTR, but without an initiation codon (ATG). Sequence of the 5-end of I.M.A.G.E. clone 5033231 is 207 bps upstream of the 5-end of I.M.A.G.E. clone 3495226, whereas its 3-end is 139 bps upstream of 3-end of I.M.A.G.E. clone 3495226. A composite 1364 bps nucleotide sequence between I.M.A.G.E. clones 3495226 and 5033231 contained two putative initiation codons, an ORF encoding a 205 amino-acid peptide with high similarity (79%) to the human TUSC1 peptide (Figure 3b). Less similarity was found between nucleotide sequences in the UTRs (data not shown). The entire cDNA sequence was found in the sequence of a single BAC clone, RP23-139B14, on mouse chromosome 4. As with the human homologue, the cDNA sequences were contiguous with the genomic sequence. The mouse homologue of TUSC1 was referred to as Tusc1 by the Mouse Genomic Nomenclature Committee (MGNC) and was deposited in GenBank (Accession no.: AY546089).
Deletion of TUSC1 in tumor DNA
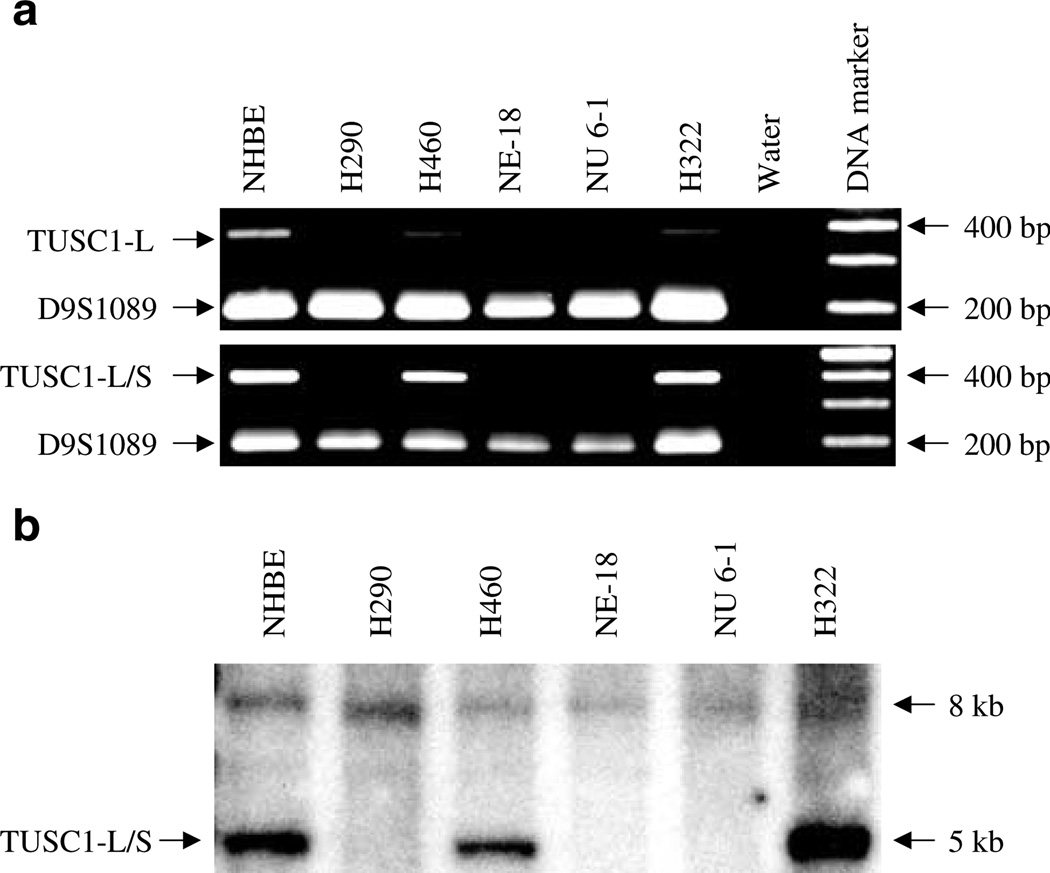
Amplification of TUSC1-L was performed with genomic DNA to study the frequency of deletion in tumor cell lines. Three NSCLC cell lines (Nu 6-1, NE-18, and H290) showed HD of TUSC1-L (Figure 1a), while it was retained in the remaining cell lines. Furthermore, multiplex PCR with primer pairs of either TUSC1-L or TUSC1-S and the microsatellite marker D9S1089 (Figure 1b) was performed with the DNA from cell lines with or without TUSC1 deletions. The marker D9S1089 amplified its fragment in all DNAs; however, there were no amplified fragments of either TUSC1-L or TUSC1-S in these three cell lines (Figure 4a). The deletions were further confirmed by Southern blot using a probe of I.M.A.G.E. 5169320 spanning both TUSC1-L and TUSC1-S (Figure 4b). A 5 kb EcoRI fragment was detected in the cell lines without HD of TUSC1, but it was not detected in the cell lines NE-18, Nu 6-1, and H290. However, an 8 kb fragment was detected in all genomic DNA, but this signal is much weaker than that of TUSC1.
Figure 4.
Deletion of TUSC1 in genomic DNA from lung cancer cells. (a) Multiplex PCR demonstrates the deletion of both TUSC1-L and TUSC1-S. (b) Deletion of TUSC1 was confirmed by Southern blot with genomic DNA digested by EcoRI. Increased intensity of the hybridized band in DNA of H322 was due to the higher amount of DNA loaded (data not shown)
Expression of TUSC1 and Tusc1 in tissue and tumor cells
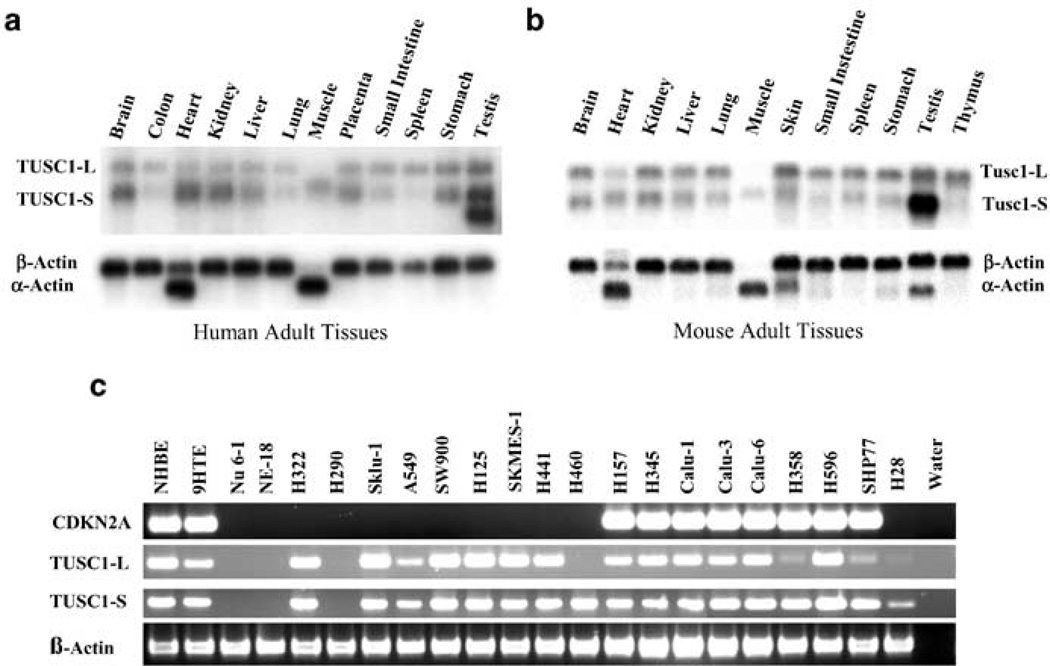
We performed Northern blot analysis with a full-length TUSC1 probe generated from clone of TUSC1-S (I.M.A.G.E. 5169320) and Tusc1 probe (I.M.A.G.E. clone 3495226) to investigate the expression pattern of TUSC1 in human and mouse adult tissues. In human tissues, Northern blot analysis revealed two major mRNA species in the majority of the tissues of approximately 2.0 and 1.5 kb. However, an additional 1.1 kb mRNA species was detected only in the testis (Figure 5a). The 2.0 kb transcript was very weak in muscle tissue, whereas the 1.5 kb transcript signals were weak in the colon, lung, and spleen tissues. All three bands of TUSC1 appeared to be stronger in the testis than in other tissues. The TUSC1-S cDNA sequence (I.M.A.G.E. 5169320) corresponds to the 1.5 kb transcript, and TUSC1-L corresponds to the 2.0 kb transcript, indicating that we have deduced two full-length cDNAs from the TUSC1 locus (Figure 3a). However, the exact source of the 1.1 kb transcript remains to be determined. A very similar expression pattern was revealed by Northern blot (Figure 5b) in mouse tissues. Two mRNA species of 2.0 and 1.5 kb with similar intensities to human tissues were detected in the majority of the tissues. Again, the intensity of the band of 1.5 kb in the testis was the strongest among signals detected the tissues (Figure 5b). As a control for RNA loading, the expected signals and intensities for β-actin or α-actin were detected in all human and mouse tissues (Figure 5a and b).
Figure 5.
Expression of TUSC1 mRNA in adult tissues and lung cancer cells. (a) Northern blot analysis of TUSC1 expression in human tissues. (b) Northern blot analysis of Tusc1 expression in mouse tissues. After hybridization to the TUSC1 or Tusc1 probe, the blot was stripped and rehybridized to a β-actin probe. (c) RT–PCR analyses for p16/CDKN2A and TUSC1 expression in lung cancer cell line mRNAs
To investigate the expression profile of TUSC1 transcripts in the tumor cell lines, we performed reverse transcriptase (RT)–PCR with specific primers for either TUSC1-L or TUSC1-S (Figure 5c). As expected, transcripts of TUSC1-L and TUSC1-S were not detected in the three NSCLC cell lines with HD (Nu 6-1, NE-18, H290; Figure 5c). Interestingly, primers specific to the 3′-end of TUSC1-L failed to amplify in the NSCLC cell line H460 (Figure 5c). Three cell lines (H358, SHP77, and H28) showed some degree of reduced product intensities compared to the other cell lines. In addition, amplification of p16/CDKN2A was performed with the same cDNA samples and expression of p16/CDKN2A was absent in 12 cell lines, including the cell lines H358, SHP77, and H28 (Figure 5c). As a positive control, a human β-actin product was amplified from all the samples (Figure 5c).
Discussion
LOH studies show that genetic alterations of chromosome 9p occur in a variety of tumor types including lung cancer, suggesting the presence of TSGs critical in the development of human cancers. Localization of HDs in tumor DNA is an important strategy to identify potential TSGs. A detailed deletion mapping study with 55 integrated STSs, microsatellite markers, and four known genes located on chromosome 9p21–24 in human lung cancer cell lines led to the identification of five regions of HDs. Of these deletions, two frequently deleted regions are clustered at the TSG p16/CDKN2A locus and the previously identified deletion at the D9S126 locus. These findings are consistent with previous reports that the p16/CDKN2A locus is a primary target for deletion in 9p. Nevertheless, additional deletions and TSGs appear to exist on chromosome 9p (Wiest et al., 1997). Interestingly, three independent novel HDs, distal to, but not contiguous with, the deletion of p16/CDKN2A, were identified in two NSCLC cell lines, NE-18 and Nu 6-1. Since the STSs and microsatellite markers used in this study are all available in the genome sequence database, these findings will provide a convenient method to analyse the genes and genome for the deleted regions described in this report. Results will be necessary and helpful to understand the involvement of HD in human carcinogenesis and how the genome rearranged chromosome 9p in the cell lines bearing the HDs.
As reported previously, both NE-18 and Nu 6-1 were clones of the human squamous lung carcinoma tumor RG-SLC-L11 (Stranahan et al., 1996). Therefore, it is not surprising to see the identical deletion patterns revealed by PCR, quantitative real-time PCR and Southern blot in these two cell lines. However, the mechanisms for the HDs in these cell lines remain to be explored. It will be useful to screen additional cell lines and primary tumors to determine if these novel deletions occur frequently. In addition, molecular cytogenetic studies such as SKY and FISH with probes from deleted regions in both cell lines may provide a clue to the cause of the chromosomal rearrangements.
The region containing D9S126 is frequently reported to be affected by HD or LOH in human cancers, for example, breast cancer, squamous cell lung cancer, melanoma, hepatocellular, suggesting that the region may contain potential TSGs. One intronless gene (TUSC1) in this region and its mouse homologue (Tusc1) were characterized in detail in this report. Two major TUSC1 transcripts, TUSC1-L and TUC1-S, were detected in a wide range of adult tissues by Northern blot analysis, indicating a ubiquitous gene expression pattern even though their expression levels differed (Figure 5a). Similar expression patterns for Tusc1 were also revealed in mouse tissues (Figure 5b). In addition, one transcript of approximately 1.1 kb was only present in human testis, and the corresponding cDNA remains to be elucidated. HD of TUSC1 in genomic DNA from the cell lines Nu 6-1, NE-18, and H290 were clearly demonstrated by multiplex PCR and in Southern blots (Figure 4a and b). However, further studies are required to determine if the weak 8 kb band detected with the TUSC1 probe represents a gene homologue of TUSC1.
RT–PCR for p16/CDKN2A and TUSC1 with cDNA from the lung tumor cell lines indicated the absence of p16/CDKN2A and TUSC1 transcripts in the cell lines with HD. It should be noted that cell line H460 lacks the expression of TUSC1 without harboring a HD. In addition, three cell lines with reduced expression of TUSC1 do not harbor an HD of the TUSC1 locus. Therefore, expression inactivation may be caused by other mechanisms, such as hypermethylation of the CpG island in the promoter region (Jones and Laird, 1999; Baylin et al., 1998; Baylin and Herman, 2000; Jones and Baylin, 2002). Finally, expression levels of TUSC1-L are reduced in two cell lines, H358 and SHP77, without apparent changes in p16/CDKN2A expression levels, suggesting that expression from these two loci are not coordinated.
Taken together, we provided strong evidence for other regions of HD on chromosome 9p in addition to the p16/CDKN2A locus in human lung cancer cells. These regions are likely to harbor potential TSGs important for the development of lung carcinogenesis. TUSC1, a novel intronless gene, was not expressed or showed reduced expression in a subset of lung cancer cells, and these findings suggest that TUSC1 may play a role as a tumor suppressor in lung tumorigenesis. Further functional and biological studies are expected to confirm if TUSC1 is a TSG and enable us to better understand the molecular mechanism of human lung cancer progression.
Materials and methods
Cell culture, RNA, and DNA extraction
The 30 NSCLC and 12 SCLC cell lines were grown in RPMI 1640 (BD Biosciences Clontech, Palo Alto, CA, USA) supplemented with 10% fetal bovine serum and 1% l-glutamine. The NSCLC cell lines were 866MT, A-2182, A-427 A549, Calu-1, Calu-3, Calu-6, ChaGo-K1, EKVX, H125, H157, H28, H290, H322, H358, H441, H460, HOP 62, HOP 92, LXFL 529, NCI-H23, NCI-H226, NCI-H292, NCI-H522, NCI-H1155, Sklu-1, SKMES-1, SW900, and two cell clones, NE-18 and Nu 6-1, derived from a human squamous lung carcinoma tumor RG-SLC-L11 (Stranahan et al., 1996). NE-18 was reported as a benign variant of Nu 6-1. The SCLC cell lines were DMS-92, DMS-114, H345, H740, H792, N417, NCI-H82, NCI-H146, NCI-H446, NCI-H526, NCI-N417, and SHP77. An NHBE was obtained and maintained following the supplier’s instructions (BioWhittaker, Walkersville, MD, USA).
Total RNA was isolated using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) following the supplier’s protocol. Extraction of DNA was performed according to a previously described protocol (Laird et al., 1991).
HD mapping by PCR screening and quantitative real-time PCR analysis
All STS and microsatellite markers used were listed in Figure 1b. HDs were initially analysed by PCR amplification with 40 STS or microsatellite markers on all 42 cell lines. In all, 15 additional markers located in or around the boundary of the identified HD were selected for further analysis with the cell lines NE-18 and Nu 6-1 shown in Figure 1b. Information for markers was obtained from the Ensembl Genome Data Resources (http://www.ensembl.org/), the Whitehead Institute (http://www-genome.wi.mit.edu/) and the National Center for Biotechnology Information (NCBI, http://ncbi.nlm.nih.gov) databases. Two primer pairs, p16/CDKN2A-F (5′-ACAAGC TTCCTTTCCGTCATGCCG-3′)/p16/CDKN2A-R (5′-TGA GCTTTGGAAGCTCTCAG-3′) and p16/CDKN2B-F (5′-TC TTTAAATGGCTCCACCTGCCTT-3′)/p16/CDKN2B-R (5′-TCCCCGTTGGCAGCCTTCATCGAATTA-3′), were used to amplify exon 2 of p16/CDKN2A and p16/CDKN2B. Two primer pairs generated from the genes INF-1 (GenBank Accession no.: X58822), INF-1F (5′-GGAGATGGTAAAAGGGAGCC-3′)/INF-1R (5′-TGCAGTTGCTGATGAAGTCC-3′), and MTAP (GenBank Accession no.: U22233) and MTAP-F (5′-CCCTTGCAATTGGAGGTAAA-3′)/MTAP-R (5′-TTATCCCAGTATCCCTCCCC-3′), were also used for HD analyses. Primers were purchased from Research Genetics (www.resgen.com) or Operon (www.operon.com). Amplification for all the markers was carried out in a 10 µl reaction mixture containing 20 ng of genomic DNA, 7.5 pmol of each primer, 2 mm of each dNTP, 1.5 mm MgCl2, and 0.25 U of Taq polymerase (Promega, Madison, WI, USA). Amplifications were performed at cycling conditions of 45 s at 95°C, 45 s at 57°C, and 60 s at 72°C for 30 cycles; the first denaturing cycle was carried out for 2 min at 95°C and the final extension was for 7 min at 72°C. Amplification for gene specific primers were 50 s at 94°C, 40 s at 58°C–60°C, and 50 s at 72°C for 30 cycles; the first denaturing cycle was carried out for 3 min at 94°C and the final extension for 3 min at 72°C. All products were separated on 2–3% agarose gels and visualized by ethidium bromide staining for analysis.
Novel HDs in the two variant cell lines, NE-18 and Nu 6-1, defined by PCR were confirmed by quantitative real-time PCR analysis on the ABI Prism 7900 Sequence Detection System (PE Applied Biosystems, Foster City, CA, USA) following SYBR Green 1 Assay Protocol provided by the supplier. The glyceraldehyde-3-phosphate dehydrogenase primer set (Mu-GAPDH, Applied Biosystems) was used as the reference control for normalization. DNA of an NHBE was used as a normal control, whereas deionized water was used as a negative control. These reactions were performed in the Gene Expression Laboratory (NCI-Frederick, MD, USA). Each sample was run in duplicate in separate wells for microsatellite and MuGAPDH. Amplification conditions were 95°C for 15 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The threshold cycles (CT) were recorded for all samples for both the microsatellite and the reference. The ΔCT value for each microsatellite was determined by subtracting the mean CT of MuGAPDH from the mean CT of the microsatellite. The ΔCT from DNA of lung cancer cell lines, NE-18 and Nu 6-1, was subsequently compared with the ΔCT from the cell line NHBE. Relative copy number was derived by the method outlined in ABI User Bulletin 2 with the formulas:
ΔCT (sample) = CT (microsatellites) − CT (MuGAPDH)
ΔΔCT = CT (tumor) − ΔCT (control)
Relative copy number = 2ΔΔCT
Isolation of human TUSC1 and mouse Tusc1
Online databases (NCBI, Ensembl Genome Data Resources, the UCSC Genome Browser: http://genome.cse.ucsc.edu/) and the computer program GENESCAN (http://genes.mit.edu/GENSCAN.html) were used for analysing the known/unknown gene(s) or exon sequences within HDs at D9S126 (Wiest et al., 1997). To establish the full-length ORFs of potential gene(s), we searched the Gene Connection™ Discovery Clones database (Stratagene, La Jolla, CA, USA) or the EST database at NCBI. Clones were purchased from Research Genetics or Stratagene and sequenced. Nucleotide sequencing for clones and PCR products cloned into pGEM-T Easy Vector (Promega, Madison, WI, USA) was performed with the PRISM BigDye Terminator Cycle Sequencing kit on an ABI 377 automated sequencer (Applied Biosystems, Foster, CA, USA). HD of TUSC1 was verified by multiplex PCR with STS marker, D9S1089, together with primer pairs from TUSC1-L (sense: 5′-AGCTTCATGTAACAGTGATCTT-3′;antisense: 5′-TTCCCCTGAAATGTTTTCCA-3′) and TUSC1-S (sense: 5′-GAAGCTGGAAGCCATGTACC-3′; antisense: 5′-AAGAGGCAGGGGAACTGTACATG-3′) and Southern blot analysis.
Northern blot and Southern blot analyses
Human and mouse 12-tissue poly-A+ RNA filters were purchased from Origene Technologies (Rockville, MD, USA) for Northern blot analysis. Genomic DNAs (10 µg) from normal and lung tumor cell lines were digested with EcoRI (New England Biolabs, Beverly, MA, USA), separated on 0.9% agarose gels, and blotted onto Hybond-N+ membranes (Amersham, Piscataway, NJ, USA) by GENIE electrophoretic blotter (Research Products International Corp, Mount Prospect, IL, USA) for Southern blot analysis. The complete insert of human I.M.A.G.E. clone 5169320 or mouse I.M.A.G.E. clone 3495226 was labeled with [α-32P]dCTP (Amersham, Piscataway, NJ, USA) using Ready-To-Go™ DNA Labeling Beads (Amersham, Piscataway, NJ, USA). Both RNA and DNA blots were hybridized at 63°C and analysed according a previously described protocol (Shan et al., 2003). The blots were stripped and rehybridized to a β-actin probe under the same conditions for sample loading controls.
RT–PCR
For RT–PCR, 2 µg of total RNA were reverse transcribed using oligo-(dT)12–18 primer and the SuperScript II Kit (Invitrogen, Carlsbad, CA, USA). Genomic DNA contamination was excluded by both DNaseI digestion of the RNA sample prior to first-strand synthesis and omission of reverse transcriptase in the reaction mixture. Amplification of the control RNA without reverse transcription did not generate any products (data not shown). Aliquots (1 µl) of the first-strand reactions were used in 25 µl amplifications with primer pairs of either TUSC1-L or TUSC1-S, described above. Since both TUSC1-L and TUSC1-S do not contain introns, the primers were also used for amplifications from genomic DNA. In addition, one sense primer, TUSC1-S1 (5′-CATGTACAGTTCCCCTGCCTCTT-3′), was used with an antisense primer, TUSC1-L1 (5′-TTCCCCTGAAATGTTTTCCA-3′), to amplify the fragment for sequence analysis of cDNA from the NHBE cell line. Amplification conditions were 50 s at 94°C, 40 s at 60°C, and 50 s at 72°C for 30 cycles; the first denaturation cycle was for 3 min at 94°C and the final extension for 3 min at 72°C. As a positive control, human β-actin primers were amplified from all RNA samples tested. In addition, two primers of p16/CDKN2A (sense: 5′-GCACCAGAGGCAGTAACCAT-3′; antisense: 5′-CATTTACGGTAGTGGGGGAA-3′) were used for RT–PCR to analyse the expression of p16/CDKN2A in the same cell lines. A measure of 15 µl of each reaction was resolved on a 2% agarose gel and visualized by ethidium bromide staining.
Acknowledgements
We would like to thank both Dr Curtis C Harris (Laboratory of Human Carcinogenesis, NCI) and the Developmental Therapeutics Program (NCI) for providing several lung cancer cell lines, Dr Douglas Lowy (Laboratory of Cellular Oncology, NCI) and CCR Fellow Editorial Board (NCI) for critical comments on the manuscript.
Footnotes
References
- An HX, Niederacher D, Picard F, van Roeyen C, Bender HG, Beckmann M. Genes Chromosomes Cancer. 1996;17:14–20. doi: 10.1002/(SICI)1098-2264(199609)17:1<14::AID-GCC3>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Adv. Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- Baylin SB, Herman JG. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- Cairns P, Polascik TJ, Eby Y, Tokino K, Califano J, Merlo A, Mao L, Herath J, Jenkins R, Westra W, Rutter JL, Buckler A, Gabrielson E, Tockman M, Cho KR, Hedrick L, Bova GS, Isaacs W, Koch W, Schwab D, Sidransky D. Nat. Genet. 1995;11:210–212. doi: 10.1038/ng1095-210. [DOI] [PubMed] [Google Scholar]
- Castellano M, Pollock PM, Walters MK, Sparrow LE, Down LM, Gabrielli BG, Parsons PG, Hayward NK. Cancer Res. 1997;57:4868–4875. [PubMed] [Google Scholar]
- Cheng JQ, Jhanwar SC, Lu YY, Testa JR. Cancer Res. 1993;53:4761–4763. [PubMed] [Google Scholar]
- Coleman A, Fountain JW, Nobori T, Olopade OI, Robertson G, Housman DE, Lugo TG. Cancer Res. 1994;54:344–348. [PubMed] [Google Scholar]
- Hamada K, Kohno T, Kawanishi M, Ohwada S, Yokota J. Cancer. 1998;22:232–240. doi: 10.1002/(sici)1098-2264(199807)22:3<232::aid-gcc9>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Hamada K, Kohno T, Takahashi M, Yamazaki M, Yamazaki M, Tashiro H, Sugawara C, Ohwada S, Sekido Y, Minna JD, Yokota J. Genes Chromosomes Cancer. 2000;27:308–318. [PubMed] [Google Scholar]
- Holland EA, Beaton SC, Edwards BG, Kefford RF, Mann GJ. Oncogene. 1994;9:1361–1365. [PubMed] [Google Scholar]
- Jones PA, Baylin SB. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Jones PA, Laird PW. Nat. Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, Johnson BE, Skolnick MH. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- Laird PW, Zijderveld A, Linders K, Rudnicki MA, Jaenisch R, Berns A. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead LJ, Gillespie MT, Hung JY, Rane US, Rayeroux KC, Irving LB, Campbell LJ. Int. J. Cancer. 1997;71:213–217. doi: 10.1002/(sici)1097-0215(19970410)71:2<213::aid-ijc15>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. Nat. Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- Oh JJ, West AR, Fishbein MC, Slamon DJ. Cancer Res. 2002;62:3207–3213. [PubMed] [Google Scholar]
- Pollock PM, Welch J, Hayward NK. Cancer Res. 2001;61:1154–1161. [PubMed] [Google Scholar]
- Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Serrano M, Hannon GJ, Beach D. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- Shan Z, Haaf T, Popescu NC. Gene. 2003;16:55–61. doi: 10.1016/s0378-1119(02)01143-5. [DOI] [PubMed] [Google Scholar]
- Sheu JC, Lin YW, Chou HC, Huang GT, Lee HS, Lin YH, Huang SY, Chen CH, Wang JT, Lee PH, Lin JT, Lu FJ, Chen DS. Br. J. Cancer. 1999;80:468–476. doi: 10.1038/sj.bjc.6690380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranahan PL, Laroe J, McCombs R, Goldsmith A, Rahim I, Overland M, Pettijohn DE. Glycoconj. J. 1996;13:741–747. doi: 10.1007/BF00702338. [DOI] [PubMed] [Google Scholar]
- Takeuchi S, Koike M, Seriu T, Bartram CR, Slater J, Park S, Miyoshi I, Koeffler HP. Leukemia. 1997;11:1636–1640. doi: 10.1038/sj.leu.2400817. [DOI] [PubMed] [Google Scholar]
- Wiest JS, Franklin WA, Otstot JT, Forbey K, Varella-Garcia M, Rao K, Drabkin H, Gemmill R, Ahrent S, Sidransky D, Saccomanno G, Fountain JW, Anderson MW. Cancer Res. 1997;57:1–6. [PubMed] [Google Scholar]
- Xiao S, Li D, Corson JM, Vijg J, Fletcher JA. Cancer Res. 1995;55:2968–2971. [PubMed] [Google Scholar]