Figure 3.
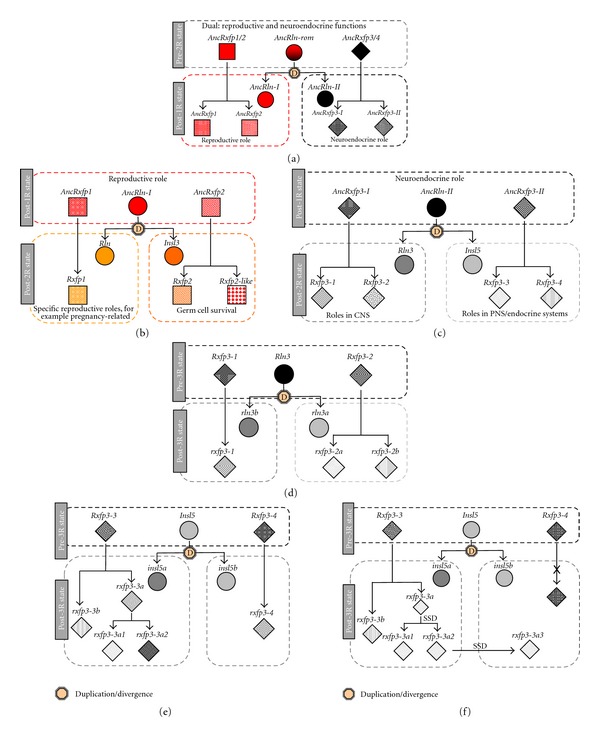
The hypothesized functional diversification of the rln/insl and rxfp genes in the gnathostome ancestors (a, b, and c) and in teleosts (d, e, and f). (a) The pre-1R three-gene system gave rise to two ligand genes and two pairs of receptor genes following 1R. After 1R, both ligands and receptors are structurally and functionally identical, which is favorable for promiscuous ligand-receptor interactions, in combination with selective pressures promoting a division of reproductive and neuroendocrine systems, leading to the establishment of novel ligand-receptor pairs. (b) Duplication and divergence of the rln-rxfp1 and insl3-rxfp2 ancestor genes. On the basis of the proposed relatedness of rxfp2-like to rxfp2, we hypothesize that Rxfp-like, at least immediately after 2R, functioned as a receptor for Insl3. (c) Duplication and divergence of the genes ancestral to rln3 and insl5 and their rxfp3/4-type receptor genes. Since all tetrapods lost rxfp3-2 and most of them also lost rxfp3-3, their ligand-receptor pairs lost their ancestral three-component nature and became two-component, that is, Rln3-Rxfp3-1 and Insl5-Rxfp4. (d) Teleosts retained all after-2R rxfp3/4 receptor genes and seem to have experienced further subfunctionalization with the formation of complex ligand-receptor relationships. We hypothesize a functional specialization of the two rln3 paralogs to work with rxfp3-1 (rln3a) and two rxfp3-2 genes. (e) Diversification of rxfp3-3 and rxfp3-4 genes in percomorpha (f) Zebrafish has lost its rxfp3-4 (i.e., rxfp4) gene but has an extra copy of rxfp3-3a3, which may imply that the receptor of Insl5b is Rxfp3-3a3. Note that in (b) and (c) insl5 paralogs are chosen arbitrarily and the interaction of the peptide with the receptors can be reversed; that is, Insl5a may function with Rxfp3-4 and Insl5b may interact with Rxfp3-3 receptors SSD = small scale duplication.
