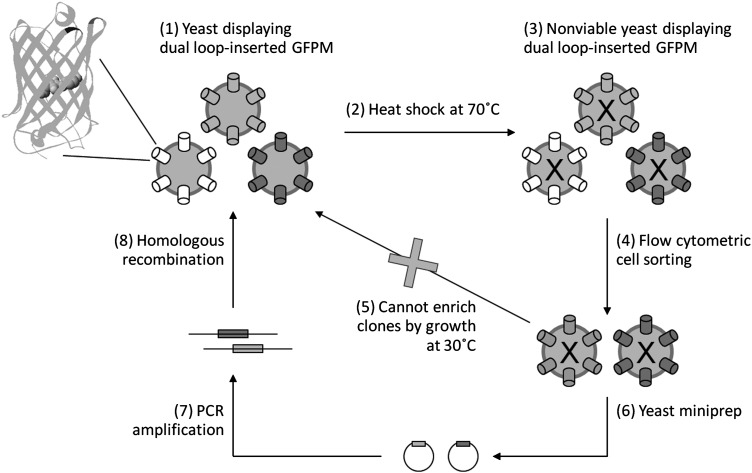
Fig. 1.
Schematic of the PCR-based procedure to engineer thermally stable proteins using yeast display. Dual-loop-inserted GFPM consists of yeast-enhanced GFP with amino acid insertions at 101–102 (black) and 172–173 (dark gray). The chromophore at center of β-barrel is shown in spacefill. Standard GFP structure pdb ID = 1EMA was used for this rendering. A library of yeast cells expressing dual-loop-inserted GFPM proteins on their surface (1) is subjected to thermal denaturation (2). The nonviable cells are then labeled with anti-c-myc antibody to measure expression, and retention of GFP fluorescence was used to monitor resistance to thermal denaturation (3). Yeast-harboring dual-loop-inserted GFPM mutants that are well expressed and resistant to thermal denaturation are recovered by flow cytometric cell sorting (4). However, the recovered yeast cells are dead and cannot be amplified by regrowth at 30°C as in traditional yeast display screens (5). Instead, plasmid DNA is recovered from the pool of sorted cells by yeast miniprep (6) and used as template for PCR-based amplification of the open reading frames of the dual-loop-inserted GFPM fragments (7). These recovered open reading frames are then used in a homologous recombination reaction to reconstruct a library enriched in thermally stable variants (8). The process is then repeated until the resultant pool is substantially enriched in dual-loop-inserted GFPM fragments that retain fluorescence after thermal denaturation.