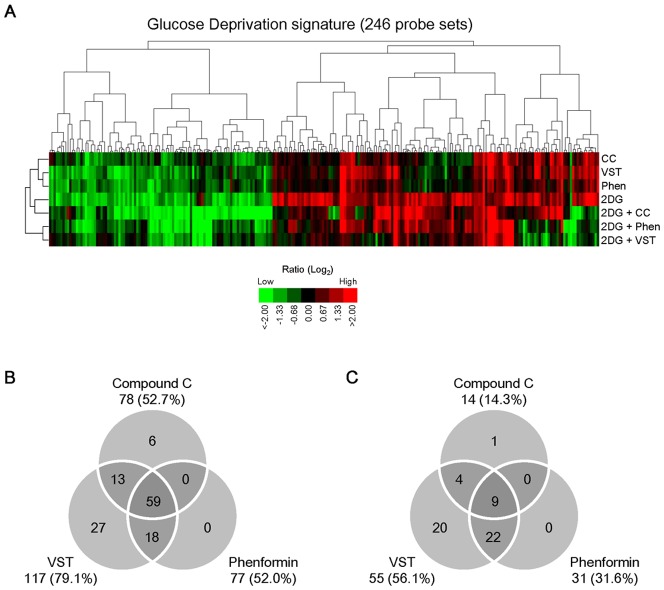
Figure 2. Inhibition of glucose deprivation-induced gene expression by compound C.
(A) Glucose deprivation signature, including 246 probe sets (X axis) sorted by cluster analysis, displayed with 7 samples (Y axis). HT1080 cells were cultured with 10 µM compound C, 10 µM versipelostatin and 100 µM phenformin for 18 h in the presence or absence of 10 mM 2DG. The log ratio for each gene was calculated by setting the expression level in appropriate control sample (non–drug-treated cells) as 0 (Log2 1). Details of the experimental conditions are provided as Table S1. 2DG, 2-deoxy-D-glucose; CC, compound C; VST, versipelostatin; Phen, phenformin. (B, C) Effects of UPR inhibitors on Glucose Deprivation signature genes. The inhibition efficiency of compound C, versipelostatin and phenformin under 2DG stress conditions was expressed using the relative fold change ratio for each of 246 probe sets, as determined by setting each 2DG-induced fold change in gene expression levels at 100%. Venn diagram shows the number of unique and shared probe sets, which indicated the inhibition efficiency ≧50% in the up-regulated 148 probe sets (B) or down-regulated 98 probe sets (C) of the Glucose Deprivation signature.