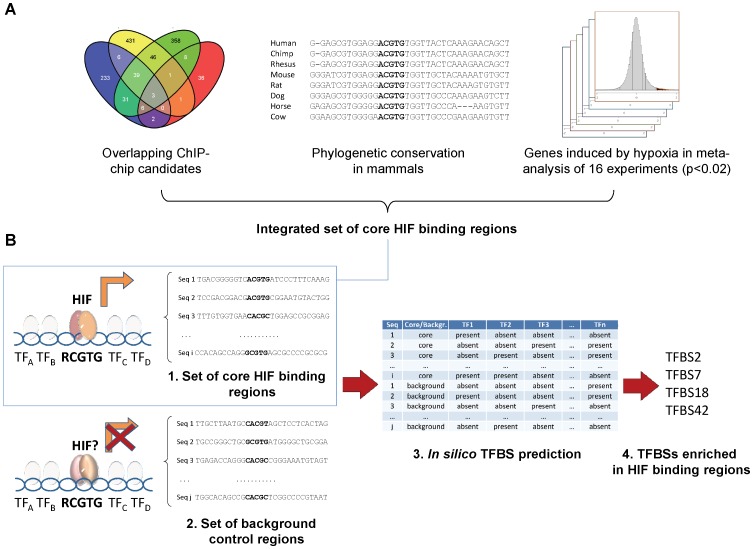
Figure 3. Integrative strategy for prediction of cooperativity in HIF binding regions.
(A) HIF1 binding locations common to at least two out of four different ChIP-chip studies in HeLa, HepG2, MCF-7 and U87 cells (left), mammalian sequence conservation of the HIF binding regions (center) and regions close to genes robustly induced in hypoxia in a meta-analysis of 16 gene expression experiments (right) were integrated into a set of bona-fide core HIF binding regions (B) Stepwise diagram for prediction and validation of TFBSs enriched in core HIF binding regions: collection of a set of core HIF binding regions and a background set of control sequences (left), in silico prediction of transcription factor binding sites present or absent in the sequences of core and background sets (center), statistical analyses of enriched TFBSs in sequences from the core set (right, top) and experimental validation of these predictions (right, bottom).