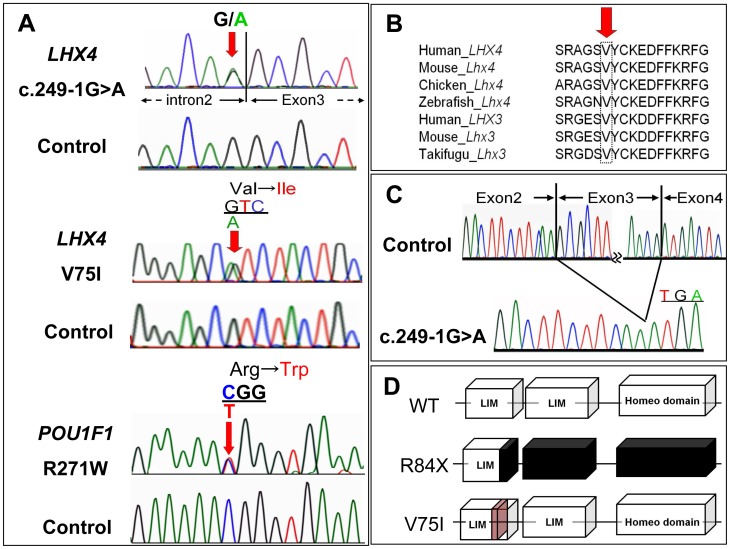
Figure 1. Identification of sequence variations of LHX4 and POU1F1.
A, Partial sequences of PCR products of the patients are shown. The upper chromatogram represents a heterozygous G to A substitution in the splice acceptor site of exon3. The middle chromatogram represents a heterozygous substitution of isoleucine (ATC) in place of valine (GTC) at codon 75. The arrow indicates the mutated nucleotide. The lower chromatogram represents a heterozygous substitution of tryptophan (TGG) in place of arginine (CGG) at codon 271. The arrow indicates the mutated nucleotide. B, Homology study showed valine at codon 75 is highly conserved through species in LHX4 and LHX3. C, Identification of exon3 skipping in the LHX4 cDNA derived from propositus of pedigree 1. LHX4 transcript with a deleted exon 3 creates a premature stop codon at the beginning of the remaining exon 4 (p.R84X). D, Schematic diagrams of the LHX4 protein. LHX4 cDNA encodes two LIM domains and one homeodomain. LHX4 with a p.R84X mutation results in the deletion of one of the two LIM domains and the entire homeodomain. Val75 is located within the first LIM domain.