Abstract
Label-free microscopy with chemical contrast and high acquisition speed up to video-rate has recently been made possible by stimulated Raman scattering (SRS) microscopy. While SRS imaging offers superb sensitivity, the spectral specificity of the original narrowband implementation is limited, making distinguishing chemical species with overlapping Raman bands difficult. Here we present a highly specific imaging method that allows mapping of a particular chemical species in the presence of interfering species based on tailored multiplex excitation of its vibrational spectrum. This is done by spectral modulation of a broadband pump beam at a high-frequency (>1MHz), allowing detection of the stimulated Raman gain signal of the narrowband Stokes beam with high sensitivity. Using the scheme, we demonstrate quantification of cholesterol in the presence of lipids, and real-time three-dimensional spectral imaging of protein, stearic acid and oleic acid in live C.elegans.
There has been increasing interest in label-free biomedical imaging based on vibrational spectroscopy. It is particularly advantageous for the imaging of small molecules, such as metabolites and drugs, because the use of fluorophores often introduces perturbations and suffers from photobleaching. Recent advances in coherent Raman scattering (CRS) microscopy, including coherent anti-Stokes Raman scattering (CARS)1-3 and stimulated Raman scattering (SRS) microscopy4-7, have led to orders of magnitude higher sensitivity than conventional Raman microscopy, and imaging speeds up to video-rate8,9.
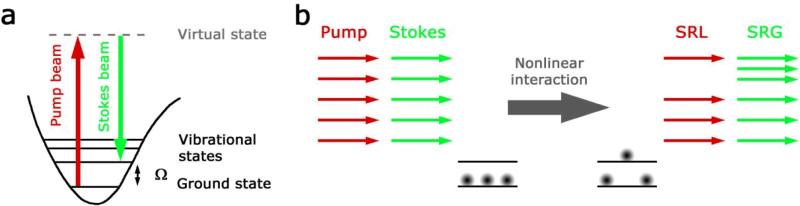
In CRS the sample is coherently excited by two lasers, one at the pump frequency, ωp, and one at the Stokes frequency, ωS. When their frequency difference Δω = ωp – ωS intrinsic vibration of the sample matches with frequency Ω, both CARS and SRS occur due to the nonlinear interaction of molecules with laser pulses (Fig. 1a). In CARS, a signal is generated at the new anti-Stokes frequency ωaS = 2ωp – ωS. In SRS, a pump photon is converted to a Stokes photon when a molecule is excited from the vibrational ground state into the first vibrational excited state. SRS thus results in an intensity loss ΔIp of the pump beam intensity Ip, and an intensity gain ΔIS of the Stokes beam intensity IS (Fig. 1b). CARS suffers from a nonresonant background signal which is present even without vibrational resonance10. SRS is free from this complication: SRS spectra are identical to those of spontaneous Raman scattering, allowing easy assignment based on Raman literature. Furthermore its sensitivity is reaching the shot-noise-limit and its signal is linear in concentration. This makes SRS more desirable than CARS for microscopy4,5,9.
Fig.1. Principle of stimulated Raman scattering (SRS).
a, Energy diagram of SRS. If the difference frequency of pump and Stokes beams is resonant with a molecular vibration, SRS occurs and a molecule of the sample is excited from its ground state to the vibrational excited state. b, As a consequence a pump photon of the excitation field is annihilated (stimulated Raman loss = SRL) and a Stokes photon is created (stimulated Raman gain = SRG) as required from energy conservation.
SRS microscopy under bio-compatible excitation conditions was recently demonstrated by implementation of a high-frequency modulation scheme to detect the relatively low SRS signal in the presence of laser noise. This also allows separation of the SRS signal from slow variations of the transmitted laser intensity due to sample scattering during raster scanning of the overlapped foci of the pump and Stokes beams5-7. To do so, either the pump or the Stokes beam is modulated at a high frequency and the modulation transfer to the other beam due to SRS of the sample is detected with phase-sensitive detection. As laser noise typically occurs at low frequencies, we use a high modulation frequency (>1MHz) to achieve near shot-noise limited sensitivity of ΔI/I < 10–8 for one second averaging time, allowing superb sensitivity in biological samples at moderate laser power5.
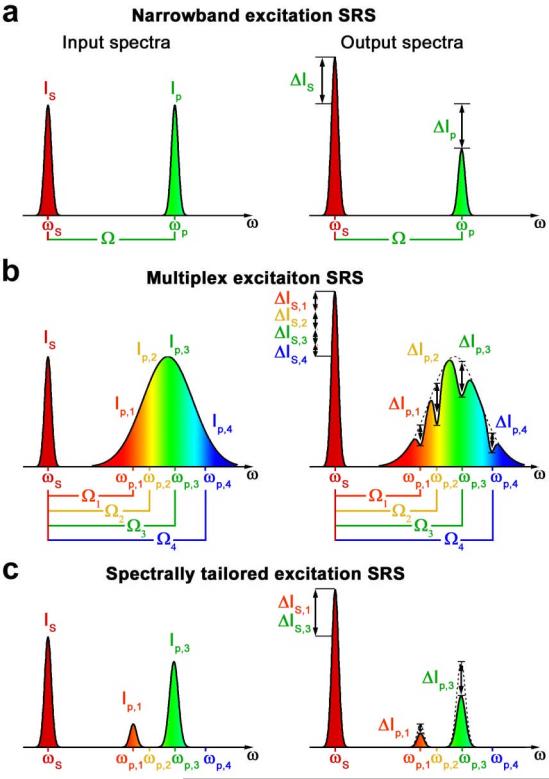
In the original implementation of SRS microscopy, we used narrowband laser beams (transform-limited picosecond pulse widths) to excite a single Raman-active vibrational mode (Fig. 2a)5. However other vibrational modes of the same species are not excited. Thus this approach does not take full advantage of the chemical specificity of Raman scattering and fails to specifically detect molecules with overlapping Raman bands.
Fig.2. Excitation schemes of SRS.
a, Narrowband excitation. Narrowband laser beams at the pump frequency ωp and Stokes frequency ωS excite a single molecular vibration Ω. The resulting SRL or SRG of the pump or Stokes intensity is measured. b, Multiplex excitation. The pump beam is broadband to excite multiple molecular vibrations Ωi of the sample simultaneously. A spectral component Ip,i with frequency ωp,i experiences an intensity loss ΔIp,i when its frequency difference with the narrowband Stokes beam Δωi = ωp,i – ωS matches Ωi. The total intensity gain of the Stokes-beam ΔIS = ΣiΔIS,i originates from the total SRS of all molecular vibrations of the sample. Micro-spectroscopy is performed by dispersing the transmitted pump light onto a multi-element detector and measuring the individual ΔIp,i. c, Spectrally tailored excitation. The broadband pump-spectrum is shaped to selectively excite only vibrations of a target molecule (e.g. Ω1 and Ω4) and avoid frequencies resonant with interfering species (e.g. Ω2 and Ω4). The total ΔIS, which only contains contributions ΔIS,1 and ΔIS,3 of the target molecule, is measured with a single-element detector.
Instead of using two narrowband pulses, we use a pump beam with a broad bandwidth and a Stokes beam with a narrow bandwidth, such that a wide spectral range of vibrational frequencies can be excited simultaneously11. Such multiplex excitation (Fig. 2b) has previously been performed in micro-spectroscopy by using an array detector and slow sample scanning4,12-14. However, such micro-spectroscopy is not compatible with high sensitivity detection because the high-frequency modulation described above cannot be employed easily. Here we present spectrally tailored excitation SRS (STESRS) microscopy, which provides images of a particular chemical species by collective excitation of selected vibrational frequencies (Fig. 2c). Such targeted excitation provides chemical selectivity based on full Raman signatures and allows fast quantification and mapping of the targeted species even in the presence of interfering species.
Principle
In STE-SRS we employ multiplex excitation with a broadband pump and a narrowband Stokes beam. The general idea is that one can tailor the pump spectrum such that it mainly probes vibrational resonances of a target species (e.g. Ω1 and Ω3 in Fig. 2c). This is done by masking the spectral components of the broadband pump pulse using a pulse-shaper with a spatial light modulator (SLM)15. We then detect the total intensity of the narrowband Stokes beam with a single photodiode, instead of multiplexed spectral detection as in micro-spectroscopy4.
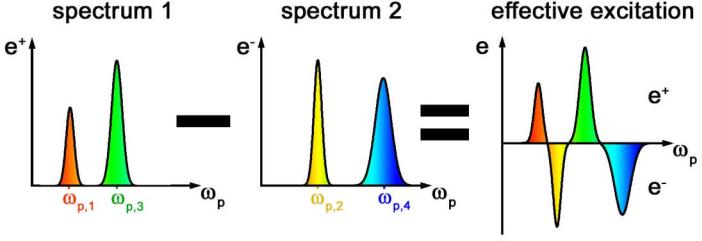
However this first excitation spectrum e+(ωp) also excites residual signal from interfering species. To discriminate against mainly probes such interference, we use another excitation spectrum e–(ωp) that mainly probes vibrational resonances of the interfering species (e.g. Ω2 and Ω4 in Fig. 3) and measure the SRS difference signal between the two excitation spectra, such that the residual signal from interfering species vanishes.
Fig. 3. Spectral modulation scheme.
While spectrally tailored excitation allows to target which vibrational resonance is excited, it does not directly improve chemical specificity compared to narrowband excitation because there is no mechanism to suppress signal from overlapping bands of interfering species. In STE-SRS, we therefore shape the broadband pump spectrum twice: The first spectrum e+(ωp) mainly contain spectral components resonant with target species (Ω1 and Ω3). The second spectrum e–(ωp) mainly contain spectral components resonant with target species (Ω2 and Ω4) and is weighted to cancel out the interfering signal generated by e+(ωp). The difference SRS signal from e+(ωp) and e–(ωp) can thus be freed from all interfering contribution. For simplicity the two spectra can be combined into one effective excitation mask e(ωp) = e+(ωp) – e–(ωp). Such subtraction can be implemented for real-time imaging by modulating between the two excitation spectra at high frequency and extracting the resulting modulation transfer with lock-in amplifier.
In a mixture of n chemical species, the contribution of a particular chemical species i to the detected SRS difference signal is linearly dependent on its concentration ci, its Raman spectrum σi(Ω = ωp – ωS and the spectrally-shaped broadband pump beam, alternating between two excitation spectra e+(ωp) and e–(ωp):
| (1) |
For simplicity, the alternating spectra can be denoted as an excitation mask e(ωp)= e+(ωp) – e–(ωp) as illustrated in Fig. 3, in which positive contributions represent e+(ωp) and the negative contributions e–(ωp).
We can now choose a particular excitation mask ej(ωp) in order to probe a target species j selectively in the presence the other species. We do so by fulfilling
| (2) |
i.e. the excitation mask ej(ωp) for species j is orthogonal to every interfering species. Hence, the contribution of each interfering species to the SRS signal vanishes independent of its concentration and the measured signal only reflects the abundance of the target species j.
In practice, the broadband pump beam can only be shaped as a collection of N discrete spectral components and the integral in (2) becomes a summation over all spectral components k
| (3) |
If the total number of species in the sample n is equal to the number of spectral components N, is uniquely determined by (3) and can be calculated by the inverse matrix of . The only inputs required are the spontaneous Raman spectra of all the species involved. Normally the total number of interfering species in a sample is much less than the N (typically 80 in our implementation) and equation system (3) is underdetermined. We therefore calculate the Moore-Penrose pseudoinverse of . This procedure is similar to the chemometric method known as classical least squares (CLS), i.e. projecting the target spectrum onto the subspace orthogonal to all interfering spectra16.
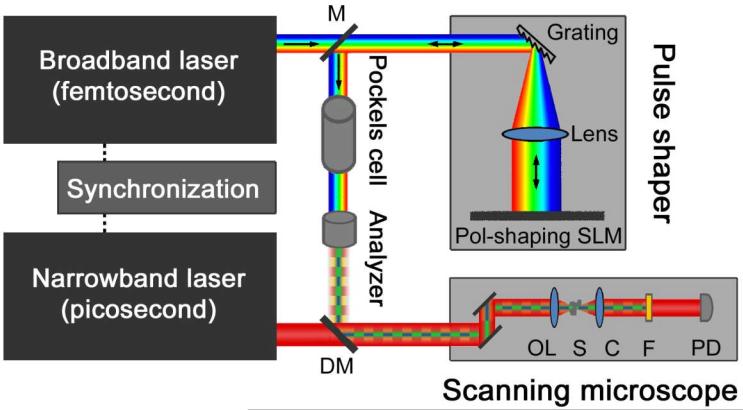
In order to detect the difference SRS signal in real-time and with high sensitivity, we modulate between the two excitation spectra at high frequency (4MHz). To do so, we combine a polarization pulse-shaper with a spatial light modulator (SLM), a polarization modulator and a polarization analyzer as shown in Fig. 4. Spectral components of the first excitation spectrum are shaped to be s-polarized and components of the second excitation spectrum are p-polarized. Fast switching between the two excitation masks is achieved by the Pockels cell. After passing through the polarization analyzer, the broadband pump beam is again linearly polarized and spectrally modulated. Different target species j can then be selected by loading different polarization masks onto the SLM in between consecutive image frames.
Fig. 4. STE-SRS microscopy setup.
Individual spectral components of the broadband pump beam are polarization-shaped in a reflection-type pulse shaper by dispersing the broadband beam with a grating onto a spatial light modulator (SLM). In- and out-going beams (arrows) are separated by a small spatial separation on a splitting mirror (M). Successive polarization modulation (Pockels cell) and passing through a polarization analyzer creates a spectrally modulated pump beam, which switches between originally s- and p-polarized spectral components. Pump and Stokes beams are spatially overlapped with a dichroic mirror (DM), temporally synchronized with electronics and aligned into a laser-scanning microscope with scanning mirrors, an objective lens (OL) and a condenser (C). After passing through the sample (S), the pump beam is blocked with an filter (F) and the Stokes beam is detected with a large-area InGaAs photo-diode (PD). The signal is analyzed with a lock-in amplifier locked into the modulation of the Pockels cell to provide the intensity of a pixel.
If such spectral modulation is performed at high frequency (higher than the low frequency laser noise), no further amplitude modulation is required for the high sensitivity detection of SRS. We measure the modulation transfer to the Stokes beam with a phase sensitivity detector (lock-in amplifier) identical to the implementation of narrowband SRS microscopy5. The amplitude of the transferred modulation is described by equation (1), as the phase-sensitive detector interprets the in and out phase signals as the first and second excitation spectra, respectively, and automatically gives the difference signal. STE-SRS has the advantage that it combines the high sensitivity of high-frequency modulation with improved spectral specificity of multiplex excitation. STE-SRS is also readily compatible with beam-scanning microscopy and allows for fast imaging speeds and real-time image display.
Results
Characterization and concentration measurement in test solutions
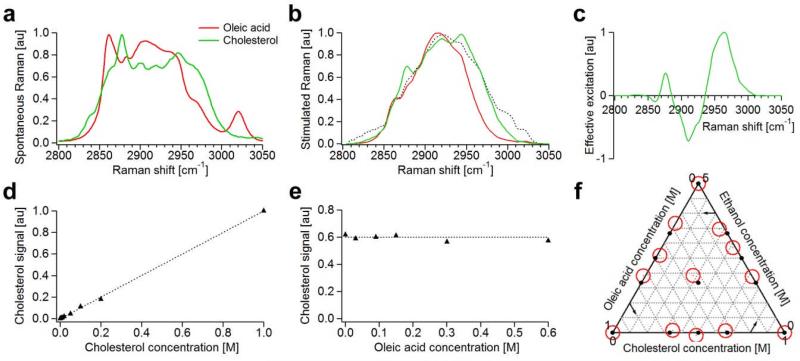
We first characterize the STE-SRS signal in a two component solutions of cholesterol and oleic acid, which have overlapping Raman bands. Fig. 5a shows the spontaneous Raman spectra of the two compounds and Fig. 5b the SRS spectra for a particular broadband pump spectrum (dotted line). We calculate the excitation mask for the detection of cholesterol (target species) in the presence of oleic acid (interfering species) from the SRS spectra as shown in Fig. 5c according to equations (3).
Fig. 5. Characterization of STE-SRS.
a, Spontaneous Raman spectra of cholesterol (green) and oleic acid (red) that have no isolated Raman vibrations but distinct Raman signatures. b, Expected SRS spectra for cholesterol (green) and oleic acid (red) calculated by normalizing the spontaneous Raman spectra with the laser excitation spectrum (black dotted). c, Excitation mask for selective detection of cholesterol (target species) in the presence of oleic acid (interfering species) generated from the SRS spectra shown in b satisfying equations (3) by the procedure described in the text. d, Linear dependence of the STE-SRS signal on concentrations of cholesterol allows for straightforward signal quantification. e, STE-SRS allows interference-free detection. STE-SRS signal with the excitation mask shown in c as a function of oleic acid concentration at constant cholesterol concentration (0.6M) dissolved in deuterated chloroform shows no false signal increase due to the increasing concentration of interferant. f, STE-SRS allows suppression against interfering species in a three component system. Ternary plot of mixtures of cholesterol, oleic acid and ethanol solutions in deuterated chloroform. Solid dots show actual concentration of the mixtures and red circles show measurement with STE-SRS.
For this excitation mask the SRS difference signal is indeed linear in the concentration of cholesterol (Fig. 5d) and independent of the concentration of oleic acid (Fig. 5e). STE-SRS can suppress against signals from interfering species that are up to ~2000× stronger (Fig. S1b), because laser intensity fluctuations of individual spectral components of a mode-locked laser are common-mode noise and can be canceled out. Such strong discrimination against interfering species is not possible with sequential measurements of different Raman bands by narrowband excitation because the signal fluctuations of different bands are uncorrelated. The detection limit of STE-SRS for cholesterol is 5mM (Fig. S1a).
We further demonstrate that STE-SRS can distinguish more than just two species. We prepare 13 different three-component solutions with varying concentrations of cholesterol, oleic acid and ethanol. For each of the solutions we sequentially apply the three excitation masks shown in supporting Fig. S2c, which are calculated for selective detection of each of three compounds in the presence of the other two based on the spontaneous Raman spectra show in Fig. S2a. We use the signal from the pure solutions to calibrate the instrument and make use of the linear concentration dependence of spectral SRS imaging to correlate the signal with the absolute concentration. The ternary plot in Fig. 5f shows that the concentration of the three-compounds can be accurately measured with STE-SRS.
Imaging proteins and specific lipids in vivo
As an important application of STE-SRS we show selective imaging of different types of fatty acids in vivo. CARS microscopy has successfully been applied to study lipid storage in the nematode C.elegans, a common model organism in lipid research17, to overcome the shortcomings of lipid staining techniques that often fail to stain all lipids uniformly18. We recently applied narrowband SRS imaging to image the distribution of unsaturated lipids in cells based on the characteristic CH-vibration5. However many lipid species, do not have isolated vibrational bands. Thus, their distributions and dynamics cannot be probed by narrowband SRS.
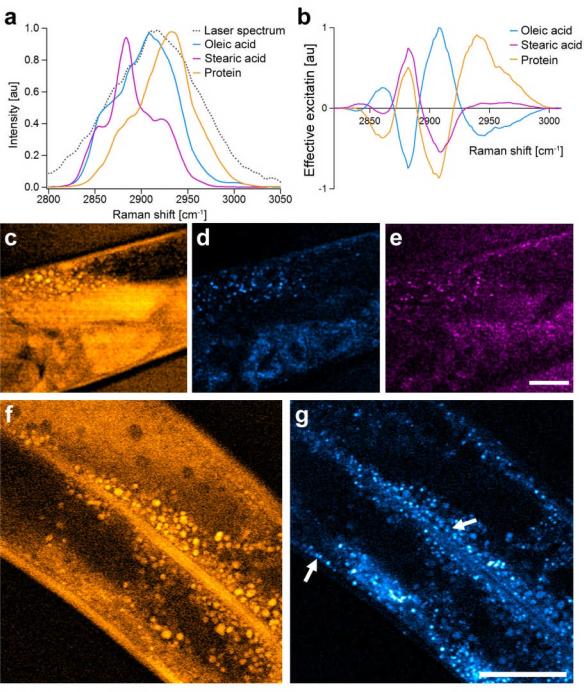
Here we image the distributions of saturated and unsaturated fatty acids as well as proteins in C.elegans, which are the three main contributors to signal in the CH-region of Raman spectra of cells18,19. Oleic and strearic acid were chosen as representative spectroscopic samples for unsaturated and saturated lipids, as they naturally occur in liquid and gel phase at room temperature. From the SRS spectra of the three pure compounds (Fig. 6a) we were able to generate three independent masks (Fig. 6b) that can selectively probe either of the three species in the presence of the two others. We can thus image a particular species selectively by applying the corresponding mask to the SLM prior to image acquisition. Before imaging the actual worm sample, we first confirmed the correct choice of the excitation masks in a test sample (supporting Fig. S3). We then took three images of the same region in the worm, one for each excitation mask, showing the distribution of protein (Fig. 6c), oleic acid (Fig. 6d) and stearic acid (Fig. 6e).
Fig. 6. Imaging of lipid storage in C.elegans.
a, SRS spectra of oleic acid (cyan), stearic acid (magenta) and protein (orange) as computed from the spontaneous Raman library by normalizing with the measured laser excitation spectrum (dotted line). b, Spectral masks computed from spectra in a and used for the imaging. c to e, Spectral images taken from the same area of a C.elegans applying spectral masks for protein (c), oleic acid (d), and stearic acid (e). Comparison of d and e shows that oleic and stearic acid deposits co-localize and that there are no isolated deposits of either one of the species. Comparison of c and d shows that the lipid deposits further co-localize with protein-dense organelles. f and g, spectral images with protein and oleic acid mask, respectively, from a different region in the worm further investigate this aspect. Arrows highlight both the sub-dermal and intestinal lipid storage depots. Imaging speed, 30s per frame with 512×512 sampling. Scalebars, 25μm.
The comparison of Fig. 6d and e shows that both compounds and their derivatives co-localize. In particular, there are no isolated storage areas that contain a single species only. Furthermore, fat deposits co-localize with areas of increased protein aggregation. Fig. 6f and g and the three-dimensional image stacks (supporting Video 1) from a different region in the worm show this in more detail. The arrows in the images indicate the two independent lipid storage areas, the subdermal and intestinal deposits. It is known that in the intestinal cells, fat is stored in lysosomal-related organelles,17 which suggests why fat deposits are surrounded by the larger protein aggregates. With STE-SRS we can further see that the fat deposits of the subdermis are also surrounded by proteins aggregates.
Discussion
Careful analyses of spectroscopic data to extract signal with a known spectral feature from complex and often noisy spectra is widespread and referred to as chemometrics20. In spontaneous Raman spectroscopy, chemometric methods have been used for understanding hydrogen bonding21, detection of glucose levels22 and bone degeneration23, and imaging of cells24. While these methods were used to analyze spectra computationally after data acquisition with a multi-element detector, recently multivariate optical computation employed a tailored multiband color filter in front of a single-element detector25,26. STE-SRS shares the same spirit, but tailors the excitation rather than emission spectra.
With STE-SRS we have introduced a microcopy technique that combines fast speed with high chemical selectivity. Micro-spectroscopy based on spontaneous Raman has been advocated recently by several groups to compliment rapid narrow band CARS imaging in order to provide spectroscopic analyses at selected positions19,27. Our spectral SRS imaging provides spectroscopic information based on multiple Raman band for every pixel of an image rather than a few selected points.
Compared to multiplex CARS microscopy12-14,28, spectral SRS imaging has the advantage that no data processing is needed as SRS excitation spectra are identical to those of spontaneous Raman scattering5,28. Furthermore, pixel dwell times in spectral SRS imaging can be orders of magnitude shorter than that in multiplex CARS14. Compared to CARS pulse-shaping approaches29-33, STE-SRS requires amplitude shaping instead of phase-shaping. Any phase manipulation, including those used in interferometric CARS34,35 are prone to phase errors in biological samples, where refractive indices vary across the sample. In general all the CARS related techniques are complicated by the coherent image artifacts due to constructive and destructive spatial interferences at the dimension comparable to the diffraction limit spot36.
We note that if unknown chemical species exist in a sample, their Raman spectra need to be taken before STE-SRS microscopy can be carried out. STE-SRS does not diminish the need for Raman micro-spectroscopy. It however significantly increases the speed of mapping chemical species, which are often known a priori in biological samples. The number of species can be in principle as high as the number of spectral pixels of tailored excitation. We also note the principle of spectrally tailored excitation can be further extended to other modulation transfer techniques such as two-color two-photon absorption37, pump-probe38, or stimulated emission39.
The combination of high specificity and high sensitivity of STE-SRS microscopy offers new prospect for vibrational imaging of for biology and medicine.
Methods
The 1064nm narrowband Stokes beam is provided from a picosecond Nd:YVO4 laser (HighQ, Picotrain) and the tunable broadband pump is obtained from a femtosecond Ti:Sa laser (Coherent, Mira900) with 76MHz repetition rate. The two lasers were synchronized using electronic synchronization (Coherent, Synchrolock) with <250fs timing jitter13,40. In order to minimize nonlinear photodamage41-43, the pump beam was chirped to picoseconds by passing the beam through 35cm of glass (Edmund Optics, NT63-091) without signal loss. Polarization pulse shaping was achieved with a custom pulse-shaper from Biophotonic Solutions Inc. The pulse-shaper consists of a grating to disperse the broadband pulse and a curved mirror (~1m focal length to achieve 0.1nm spectral resolution) to focus the spectral components onto a polarization spatial light modulator (CRI, SLM-640) in a 4-f-geometry in reflection mode15. The through-put of the pulse-shaper was ~55±5% at 800nm center wavelength depending on the input polarization. The SLM was controlled using custom software control using LabView (National Instruments). To allow fast modulation between different polarization states (at 4MHz) we utilized a combination of a custom-built Pockels cell (based on RTP crystals by Raicol) and polarization analyzer. The Pockels cell was driven with a sine-wave from the reference output from the lock-in amplifier (Stanford Research Systems, SR844RF), which was amplified to 1W and efficiently coupled to the crystal with a resonant transformer. Pump and Stokes beams were spatially overlapped using an 850nm long-pass mirror (Chroma Technology), temporally overlapped with the synchronization electronics by maximizing the cross-correlation signal on an autocorrelator (Spectra-Physics, 409 Autocorrelator) and coupled into a modified laser-scanning upright microscope (Olympus, BX61WI/FV300). We used a 60× 1.2NA water (Olympus, UPlanApo / IR) as excitation objective and the light is collected in transmission with a 1.4NA oil condenser (Nikon). For imaging of biological samples, the average power was reduced to 15mW of pump and 120mW of Stokes. A large-area InGaAs photodiode (New England Photoconductors, I5-3-5) with reversed bias of 12V is used for detection of the narrowband Stokes beam, after blocking the spectrally modulated pump beam with a high OD long-pass filter (Chroma Technology, HHQ925LP). The photodiode output is band-pass-filtered around the modulation rate of 4MHz with a custom-made bandpass filter. A high-frequency lock-in amplifier (Stanford Research Systems, SR844RF) is used to demodulate the Stokes-intensity. Time-constant of 10μs (“no filter”-mode) was used for imaging and 1s for the solution spectroscopy studies. The analog output of the lock-in amplifier is fed into a input of the microscope A/D-converter to provide the intensity of a pixel.
Supplementary Material
Acknowledgements
The authors thank Linjiao Luo and Aravinthan Samuel for providing the C.elegans sample for initial testing, Brian Saar and Sijia Lu for helpful discussions and commenting on the manuscript and Xu Zhang for assisting in the final measurements. C.W.F. acknowledges Boehringer Ingelheim Fonds for a Ph.D. Fellowship. This work was supported by the NIH Director's Pioneer Award and NIH TR01 grant 1R01EB010244-01.
Footnotes
Author contributions
C.W.F., W.M. and X.S.X. conceived the idea and drafted the manuscript. C.W.F. and G.R.H. built the instrument, B.X. and M.D. designed and built the pulse shaper, and C.W.F. conducted the experiments.
Additional information
A patent application based on this work has been filed. Supplementary information accompanies this paper at www.nature.com/naturephotonics.
Bibliography
- 1.Zumbusch A, Holtom GR, Xie XS. Three-dimensional vibrational imaging by coherent anti-Stokes Raman scattering. Physical Review Letters. 1999;82:4142–4145. [Google Scholar]
- 2.Cheng JX, Xie XS. Coherent anti-Stokes Raman scattering microscopy: Instrumentation, theory, and applications. Journal of Physical Chemistry B. 2004;108:827–840. [Google Scholar]
- 3.Evans CL, Xie XS. Coherent anti-Stokes Raman scattering microscopy: Chemical imaging for biology and medicine. Annual Review of Analytical Chemistry. 2008;1:27. doi: 10.1146/annurev.anchem.1.031207.112754. [DOI] [PubMed] [Google Scholar]
- 4.Ploetz E, Laimgruber S, Berner S, Zinth W, Gilch P. Femtosecond stimulated Raman microscopy. Applied Physics B. 2007;87:389–393. [Google Scholar]
- 5.Freudiger CW, et al. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science. 2008;322:1857–1861. doi: 10.1126/science.1165758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozeki Y, Dake F, Kajiyama S, Fukui K, Itoh K. Analysis and experimental assessment of the sensitivity of stimulated Raman scattering microscopy. Opt Express. 2009;17:3651–3658. doi: 10.1364/oe.17.003651. [DOI] [PubMed] [Google Scholar]
- 7.Nandakumar P, Kovalev A, Volkmer A. Vibrational imaging based on stimulated Raman scattering microscopy. New Journal of Physics. 2009;11 [Google Scholar]
- 8.Evans CL, et al. Chemical imaging of tissue in vivo with video-rate coherent anti-Stokes Raman scattering microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16807–16812. doi: 10.1073/pnas.0508282102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saar BG, et al. Video-Rate Molecular Imaging In Vivo with Stimulated Raman Scattering. Science. 2010 doi: 10.1126/science.1197236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levenson MD, Kano SS. Introduction to Nonlinear Laser Spectroscopy. Academic Press Inc.; 1988. [Google Scholar]
- 11.Kukura P, McCamant DW, Mathies RA. Femtosecond stimulated Raman spectroscopy. Annual Review of Physical Chemistry. 2007;58:461–488. doi: 10.1146/annurev.physchem.58.032806.104456. [DOI] [PubMed] [Google Scholar]
- 12.Wurpel GWH, Schins JM, Muller M. Chemical specificity in three-dimensional imaging with multiplex coherent anti-Stokes Raman scattering microscopy. Opt Lett. 2002;27:1093–1095. doi: 10.1364/ol.27.001093. [DOI] [PubMed] [Google Scholar]
- 13.Cheng JX, Volkmer A, Book LD, Xie XS. Multiplex coherent anti-Stokes Raman scattering microspectroscopy and study of lipid vesicles. Journal of Physical Chemistry B. 2002;106:8493–8498. [Google Scholar]
- 14.Rinia HA, Burger KNJ, Bonn M, Muller M. Quantitative label-free imaging of lipid composition and packing of individual cellular lipid droplets using multiplex CARS microscopy. Biophys J. 2008;95:4908–4914. doi: 10.1529/biophysj.108.137737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiner AM. Femtosecond pulse shaping using spatial light modulators. Rev Sci Instrum. 2000;71:1929–1960. [Google Scholar]
- 16.Wise BM, et al. PLS_Toolbox 4.0 - Manual. Eigenvector Research; 2006. [Google Scholar]
- 17.Mullaney BC, Ashrafi K. C.elegans fat storage and metabolic regulation. Biochimica Et Biophysica Acta-Molecular and Cell Biology of Lipids. 2009;1791:474–478. doi: 10.1016/j.bbalip.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hellerer T, et al. Monitoring of lipid storage in Caenorhabditis elegans using coherent anti-Stokes Raman scattering (CARS) microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14658–14663. doi: 10.1073/pnas.0703594104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slipchenko MN, Le TT, Chen HT, Cheng JX. High-speed vibrational imaging and spectral analysis of lipid bodies by compound Raman microscopy. Journal of Physical Chemistry B. 2009;113:7681–7686. doi: 10.1021/jp902231y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mark H, Workman J. Chemometrics in Spectroscopy. Adcademic Press; 2007. [Google Scholar]
- 21.Perera PN, et al. Observation of water dangling OH bonds around dissolved nonpolar groups. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12230–12234. doi: 10.1073/pnas.0903675106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enejder AMK, et al. Raman spectroscopy for noninvasive glucose measurements. J Biomed Opt. 2005;10 doi: 10.1117/1.1920212. [DOI] [PubMed] [Google Scholar]
- 23.Schulmerich MV, et al. Noninvasive Raman tomographic imaging of canine bone tissue. J Biomed Opt. 2008;13 doi: 10.1117/1.2904940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pully VV, Lenferink A, Otto C. Raman-fluorescence hybrid microspectroscopy of cell nuclei. Vibrational Spectroscopy. 2010;53:12–18. [Google Scholar]
- 25.Nelson MP, Aust JF, Dobrowolski JA, Verly PG, Myrick ML. Multivariate optical computation for predictive spectroscopy. Analytical Chemistry. 1998;70:73–82. doi: 10.1021/ac970791w. [DOI] [PubMed] [Google Scholar]
- 26.Uzunbajakava N, de Peinder P, 't Hooft GW, van Gogh ATM. Low-cost spectroscopy with a variable multivariate optical element. Analytical Chemistry. 2006;78:7302–7308. doi: 10.1021/ac060985o. [DOI] [PubMed] [Google Scholar]
- 27.Krafft C, et al. A comparative Raman and CARS imaging study of colon tissue. Journal of Biophotonics. 2009;2:303–312. doi: 10.1002/jbio.200810063. [DOI] [PubMed] [Google Scholar]
- 28.Rinia HA, Bonn M, Muller M. Quantitative multiplex CARS spectroscopy in congested spectral regions. Journal of Physical Chemistry B. 2006;110:4472–4479. doi: 10.1021/jp0564849. [DOI] [PubMed] [Google Scholar]
- 29.Dudovich N, Oron D, Silberberg Y. Single-pulse coherently controlled nonlinear Raman spectroscopy and microscopy. Nature. 2002;418:512–514. doi: 10.1038/nature00933. [DOI] [PubMed] [Google Scholar]
- 30.van Rhijn ACW, Postma S, Korterik JP, Herek JL, Offerhaus HL. Chemically selective imaging by spectral phase shaping for broadband CARS around 3000 cm-1. J Opt Soc Am B. 2009;26:559–563. [Google Scholar]
- 31.Marks DL, Geddes JB, Boppart SA. Molecular identification by generating coherence between molecular normal modes using stimulated Raman scattering. Opt Lett. 2009;34:1756–1758. doi: 10.1364/ol.34.001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oron D, Dudovich N, Silberberg Y. All-optical processing in coherent nonlinear spectroscopy. Physical Review A. 2004;70 [Google Scholar]
- 33.Roy S, Wrzesinski P, Pestov D, Dantus M, Gord JR. Single-beam coherent anti-Stokes Raman scattering (CARS) spectroscopy of gas-phase CO2 via phase and polarization shaping of a broadband continuum. Journal of Raman Spectroscopy. 2010 [Google Scholar]
- 34.Evans CL, Potma EO, Xie XSN. Coherent anti-Stokes Raman scattering spectral interferometry: determination of the real and imaginary components of nonlinear susceptibility chi((3)) for vibrational microscopy. Opt Lett. 2004;29:2923–2925. doi: 10.1364/ol.29.002923. [DOI] [PubMed] [Google Scholar]
- 35.Jurna M, Korterik JP, Otto C, Herek JL, Offerhaus HL. Background free CARS imaging by phase sensitive heterodyne CARS. Opt Express. 2008;16:15863–15869. doi: 10.1364/oe.16.015863. [DOI] [PubMed] [Google Scholar]
- 36.Cheng JX, Xie XS. Green's function formulation for third-harmonic generation microscopy. J Opt Soc Am B. 2002;19:1604–1610. [Google Scholar]
- 37.Fu D, Ye T, Matthews TE, Yurtsever G, Warren WS. Two-color, two-photon, and excited-state absorption microscopy. J Biomed Opt. 2007;12 doi: 10.1117/1.2780173. [DOI] [PubMed] [Google Scholar]
- 38.Fu D, et al. Probing skin pigmentation changes with transient absorption imaging of eumelanin and pheomelanin. J Biomed Opt. 2008;13 doi: 10.1117/1.2976424. [DOI] [PubMed] [Google Scholar]
- 39.Min W, et al. Imaging chromophores with undetectable fluorescence by stimulated emission microscopy. Nature. 2009;461:1105–1109. doi: 10.1038/nature08438. [DOI] [PubMed] [Google Scholar]
- 40.Jones DJ, et al. Synchronization of two passively mode-locked, picosecond lasers within 20 fs for coherent anti-Stokes Raman scattering microscopy. Rev Sci Instrum. 2002;73:2843–2848. [Google Scholar]
- 41.Hopt A, Neher E. Highly nonlinear photodamage in two-photon fluorescence microscopy. Biophys J. 2001;80:2029–2036. doi: 10.1016/S0006-3495(01)76173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nan XL, Potma EO, Xie XS. Nonperturbative chemical imaging of organelle transport in living cells with coherent anti-stokes Raman scattering microscopy. Biophys J. 2006;91:728–735. doi: 10.1529/biophysj.105.074534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu Y, Wang HF, Shi RY, Cheng JX. Characterization of photodamage in coherent anti-Stokes Raman scattering microscopy. Opt Express. 2006;14:3942–3951. doi: 10.1364/oe.14.003942. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.