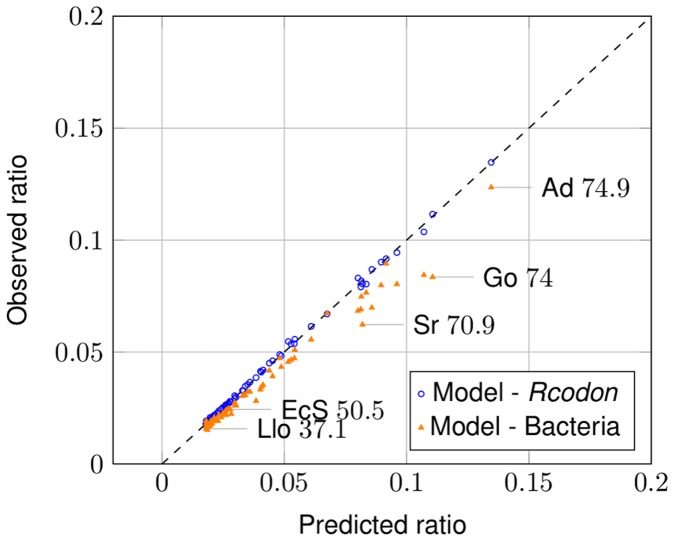
Figure 5. Ratio of annotated ORFs to non-annotated ORFs.
The ratio predicted by the mixture model is compared to the ratio observed in bacterial genomes (orange triangles) and Rcodon (blue open dots), respectively. The observable slight difference between natural genomes and Rcodon is due the fact that the expected number of short coding ORFs in Rcodon deviates from the natural genomes (compare to Figure 7). Some individual data points are labeled with an abbreviated species name and the corresponding GC-content according to Table S1.