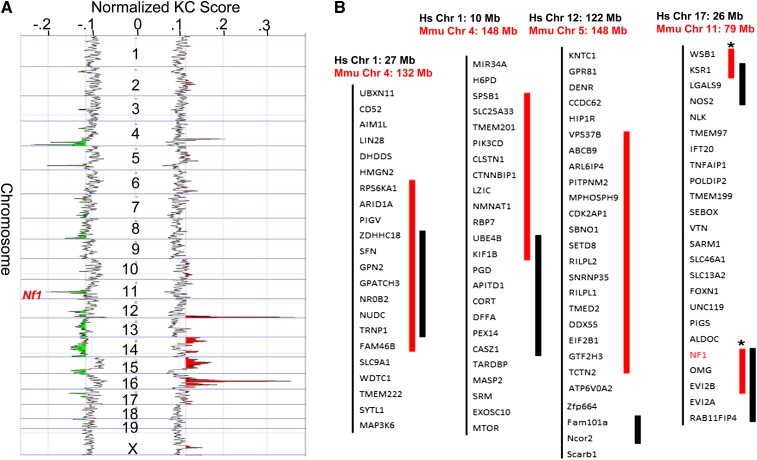
Figure 2 .
Recurrent CNAs in Chaos3 mammary tumors overlap with those in human breast cancer, including Nf1 deletion. (A) KCSmart analysis of combined aCGH data from 12 Chaos3 tumors, (nine mammary and three nonmammary). The most significant amplification peaks (red) lie on Chrs 12 and 16, and deletions (green) on Chrs 4, 5, and 11. (B) Overlap of mouse (Mmu) Chaos3 recurrent deletions with human (Hs) breast tumor CNAs. Human gene orders are shown. Thick red bars indicate the critical regions of mouse deletions, the subregions that, among all alterations in those regions, are common across all or most Chaos3 tumors. Of Chaos3 tumors with CNAs in these regions, the percentage of those containing the critical region is as follows: Chr 4 132 M = 86%; Chr 4 148 M = 71%; Chr 5 = 86%; Chr 11 = 86% (100% for Nf1, Ksr1, and Wsb1). The thick black bars to the right of the gene symbols indicate corresponding recurrent segmental CNAs in human breast cancers (limited level 4 data set from TCGA). Note that the Chr 11 deletions are single events in mice (asterisks on red part indicate these sequences are juxtaposed and contiguous in the mouse genome), and it is possible that the interval between NOS2 and NF1 may also be deleted as single events in human breast cancers, since the intervening genes are present in the hemizygous state in a high percentage of tumors according to extended TCGA data sets.