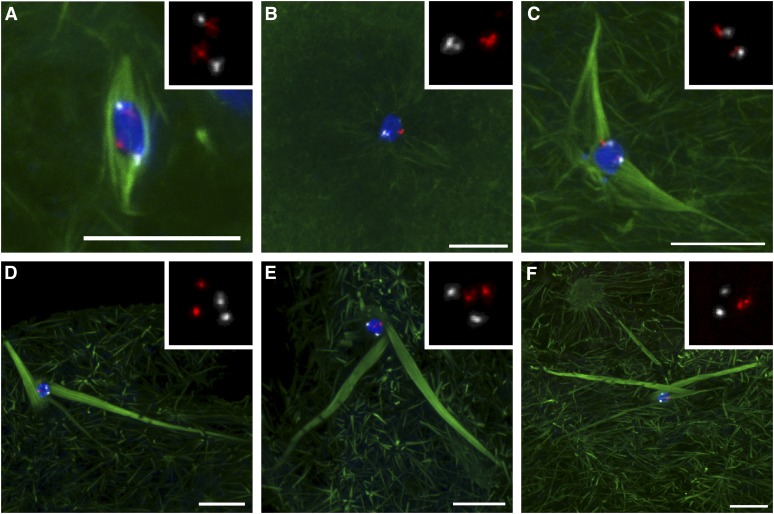
Figure 5 .
Biorientation of homologous chromosomes is defective in Klp10A germline mutants. (A) In wild type, both the 2nd (red) and 3rd (white) chromosome centromeric FISH probes show biorientation toward opposite spindle poles. All 17 centromere pairs scored were separated (two FISH signals) and oriented correctly. (B) Klp10A germline mutant oocyte prior to or during nuclear envelope breakdown. Homologous centromeres are paired. (C) Klp10A germline mutant oocyte with a long and disorganized spindle, which is nonetheless bipolar and has properly oriented centromeres. (D) Klp10A germline mutant oocyte with a disorganized spindle, loosely “bioriented” chromosomes, but “mono-oriented” 3rd chromosomes. (E and F) Klp10A germline mutant oocytes with disorganized spindles and randomly oriented centromeres. Centromeres in E are not associated with microtubule bundles. DNA is in blue and tubulin is in green. Insets show only the FISH signals. Bars, 10 μm.