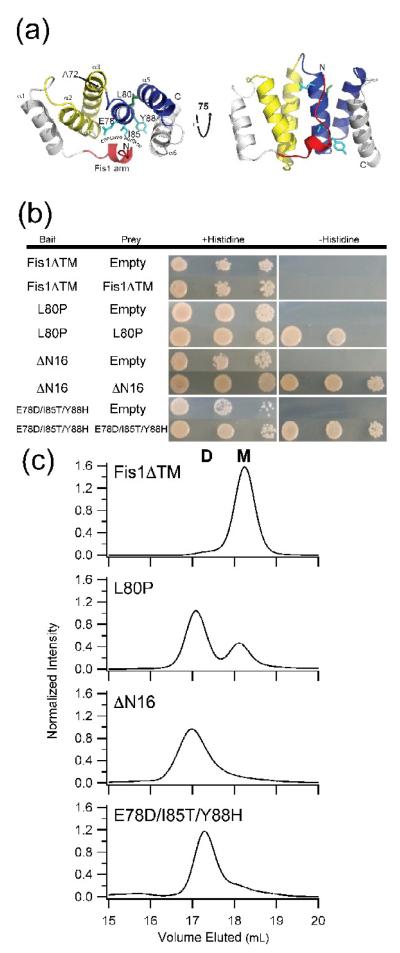
Figure 1. The cytoplasmic domains of non-functional Fis1 variants self-associate.
(a) Cartoon representation of the cytoplasmic domain of yeast Fis1 (Fis1ΔTM, 3o48.pdb). Three non-functional variants of Fis1 (L80P (green), ΔN16 (red), and Fis1ΔTM-3 (cyan, E78D/I85T/Y88H)) are highlighted by stick representation of the wild type residues. These variants have previously been reported to impair mitochondrial fission. The two TPR-like domains are colored yellow and blue. This figure was made with PyMOL86. (b) The cytoplasmic domains of non-functional Fis1 variants, but not wild type, interact by yeast two-hybrid. Yeast two-hybrid assays using the HIS3 reporter (sequential ten-fold yeast dilutions at 1, 0.1, and 0.01 OD600 are shown) with the indicated bait and prey constructs. Cells were plated onto media containing histidine (growth control) and media lacking histidine to select for yeast two-hybrid interactions and photographed after incubation for 3 days at 30 °C. (c) Fis1ΔTM variants elute by gel filtration chromatography as dimers to differing degrees. 2.5 μM of each of the indicated recombinantly expressed proteins were separated by a Superdex 200 10/300 GL at 25 °C (50 mM sodium phosphate, 184 mM NaCl, 5 mM EDTA and 2 mM DTT, pH 7.4). Each mutant is well-folded by circular dichroism spectrapolarimetry (Supplemental Figure 1).