Abstract
This study shows that naturally occurring Vibrio predatory bacteria (VPB) exert a major role in controlling pathogenic vibrios in seawater and shellfish. The growth and persistence of Vibrio parahaemolyticus and Vibrio vulnificus were assessed in natural seawater and in the Eastern oyster, Crassostrea virginica. The pathogens examined were V. vulnificus strain VV1003, V. parahaemolyticus O1:KUT (KUT stands for K untypeable), and V. parahaemolyticus O3:K6 and corresponding O3:K6 mutants deficient in the toxRS virulence regulatory gene or the rpoS alternative stress response sigma factor gene. Vibrios were selected for streptomycin resistance, which facilitated their enumeration. In natural seawater, oysters bioconcentrated each Vibrio strain for 24 h at 22°C; however, counts rapidly declined to near negligible levels by 72 h. In natural seawater with or without oysters, vibrios decreased more than 3 log units to near negligible levels within 72 h. Neither toxRS nor rpoS had a significant effect on Vibrio levels. In autoclaved seawater, V. parahaemolyticus O3:K6 counts increased 1,000-fold over 72 h. Failure of the vibrios to persist in natural seawater and oysters led to screening of the water samples for VPB on lawns of V. parahaemolyticus O3:K6 host cells. Many VPB, including Bdellovibrio and like organisms (BALOs; Bdellovibrio bacteriovorus and Bacteriovorax stolpii) and Micavibrio aeruginosavorus-like predators, were detected by plaque assay and electron microscopic analysis of plaque-purified isolates from Atlantic, Gulf Coast, and Hawaiian seawater. When V. parahaemolyticus O3:K6 was added to natural seawater containing trace amounts of VPB, Vibrio counts diminished 3 log units to nondetectable levels, while VPB increased 3 log units within 48 h. We propose a new paradigm that VPB are important modulators of pathogenic vibrios in seawater and oysters.
INTRODUCTION
Vibrio parahaemolyticus is the principal source of seafood-associated bacterial illness in the United States and the Far East (27). In 1995, routine surveillance for diarrheal diseases in Kolkata, India, revealed a high frequency of a novel V. parahaemolyticus serotype, O3:K6 (30). Its rapid spread to Asia, North and South America, Africa, and Europe was a cause for concern because high hospital admission rates suggested that this serotype might be more virulent than other strains (27). The first outbreak of V. parahaemolyticus O3:K6 in the United States was reported in 1998 from the consumption of oysters from Texas and led to 296 illnesses (8). Another outbreak occurred from oysters and clams harvested from Long Island, New York, causing illnesses in Connecticut, New Jersey, and New York (4). Even though illnesses from V. parahaemolyticus O3:K6 have been reported in the United States, the number of outbreaks is very low compared to many other countries. The reason for the limited outbreaks in the United States remains uncertain.
A variety of potential virulence factors have been identified for V. parahaemolyticus O3:K6. Studies showed the unique presence of at least four genomic islands in the pandemic V. parahaemolyticus RIMD2210633, an O3:K6 strain isolated from Japan in 1996 (2, 14). It was hypothesized that genomic island DNA may be associated with increased pathogenicity toward humans or increased fitness in the aquatic environment (14). A region named Vibrio parahaemolyticus island 7 on chromosome II contains two copies of the tdh gene, which encodes the thermostable direct hemolysin, an important virulence factor in V. parahaemolyticus (24, 29). In addition, this region encodes a type three secretion system (T3SS) named T3SS-2. Genome sequence analysis also identified the presence of a second T3SS (T3SS-1 or TTSS-1) on chromosome I (24). Both T3SSs are important for virulence, and a number of effector proteins have been identified (18). Recently, we demonstrated that the Vibrio-specific ToxR and ToxS transmembrane transcriptional regulatory proteins are important for T3SS-1 expression (42). ToxRS also protects V. parahaemolyticus from the stresses of acid, bile salts, and detergents via its positive regulation of the outer membrane protein OmpU (42, 43).
The σ38 alternative stress response gene, rpoS, encodes the RpoS protein, which exerts a role in the response of Vibrio and other bacteria to stress, including acid, cold, heat, osmotic shock, starvation, salinity, pH, and oxidative damage (13, 22, 23, 31, 35, 39, 40, 43). RpoS also upregulates many chemotaxis and motility genes (28) and a variety of virulence genes (1, 10, 12, 19). Since rpoS and toxRS are integrally involved in Vibrio survival and virulence, we hypothesized that these genes would influence the uptake or colonization of V. parahaemolyticus O3:K6 in Eastern oysters or Vibrio survival in seawater. Other serotypes of V. parahaemolyticus have been associated with outbreaks of seafood-associated illness, including a pandemic strain known as V. parahaemolyticus O1:KUT (KUT stands for K untypeable) (15). Vibrio vulnificus is another species which is of concern and has a mortality rate of more than 50% (16). It is most frequently transmitted through the consumption of raw or undercooked oysters and through wounds sustained in the marine environment.
Bdellovibrio and like organisms (BALOs) are Gram-negative bacteria that are predatory toward a host of other Gram-negative bacteria, including vibrios. They are phylogenetically and environmentally diverse (32) and have complicated life cycles, with both host-dependent and host-independent replication (3, 9, 36, 44). In host-dependent replication, small BALOs with single polar flagella, known as attack phase BALOs, encounter a susceptible Gram-negative host bacterium, enzymatically digest a hole in the host membrane, and enter the cell (38). The invader then forms an encystment, known as a bdelloplast, within the periplasmic space of the host. The bdelloplast contains a replicative form, which resembles an elongating worm that derives nutrients for growth from the host. Upon maturation, the bdelloplast septates into multiple (usually 3 to 9) immature cells. The small immature cells are released from the host and grow into mature, attack phase cells (34). The attack phase BALOs are reportedly only 0.2 to 0.4 μm in diameter (44), and the immature forms are smaller.
An alternative means of BALO replication is through the host-independent pathway, which is where BALOs replicate external to any prey or in the absence of prey. In this process, the BALOs can form chains of variable length in the extracellular milieu. Chains subsequently septate to form individual BALOs capable of reinitiating the replication process. In other cases, extracellular reproduction can occur in a manner reminiscent of typical bacteria, via binary fission into two progeny of equal sizes (9). Another type of vibrio predatory bacterium is Micavibrio aeruginosavorus, a bacterium that resembles attack phase BALOs in size and shape but that parasitizes and kills host cells without entering the host (21). They have been identified as an important factor in killing bacteria in biofilms (17).
In this paper, we do the following: (i) show a general decrease of V. parahaemolyticus O3:K6, V. parahaemolyticus O1:KUT, and V. vulnificus in natural seawater and oysters over time; (ii) demonstrate through gene knockout that toxRS and rpoS have no effect on V. parahaemolyticus O3:K6 survival or proliferation in seawater or oysters; (iii) identify the cause of the Vibrio reductions as BALOs and other Vibrio predatory bacteria (VPB); (iv) show a reduction in Vibrio levels and a concomitant increase in the levels of VPB in seawater over time; and (v) assess the levels of VPB in natural seawater obtained from various locations. This work suggests a new paradigm of the importance of VPB in modulating environmental vibrios.
MATERIALS AND METHODS
Bacterial and algal cultures.
The Vibrio parahaemolyticus and Vibrio vulnificus cultures used in this study are described in Table 1 and were derived from clinical isolates. V. parahaemolyticus O3:K6 strain RIMD2210633, isolated from an outbreak in Japan, was selected for streptomycin resistance as previously described (43) and was used as the wild-type strain. In addition, streptomycin-resistant toxRS and rpoS deletion mutants (ΔtoxRS and ΔrpoS) were constructed from the wild-type strain (43). The O1:KUT (K untypeable) serotype of another pandemic V. parahaemolyticus (15) and a highly virulent V. vulnificus (strain VV1003) are described in Table 1. High virulence of V. vulnificus VV1003 was previously determined in mouse 50% lethal dose (LD50) assays (33). These species and strains were naturally selected for streptomycin resistance. The vibrios were routinely maintained in Luria-Bertani (LB) broth or agar (Becton, Dickinson and Co., Sparks, MD) containing a total of 3% NaCl (LB–3% NaCl broth or agar) at 37°C. Stock cultures were obtained from the American Type Culture Collection (ATCC) (Manassas, VA) and consisted of a host-dependent Bdellovibrio bacteriovorus (ATCC 15143) provided as a coculture with Escherichia coli host cells (ATCC 15144), and a prey-independent strain of Bacteriovorax stolpii (ATCC 27052), which is also propagatable in E. coli B cells (ATCC 11303). Stock cultures were grown under the conditions recommended by the ATCC.
Table 1.
Vibrios used in this study
| Species | Serogroup and relevant genotypea | Strain | Source | Strb | Reference |
|---|---|---|---|---|---|
| V. parahaemolyticus | O3:K6 toxRS+ rpoS+ | Wild-type RIMD2210633 | E. F. Boyd, University of Delaware, Newark, DE | + | 43 |
| O3:K6 toxRS rpoS+ | ΔtoxRS deletion mutant | E. F. Boyd, University of Delaware | + | 43 | |
| O3:K6 toxRS+ rpoS | ΔrpoS deletion mutant | E. F. Boyd, University of Delaware | + | 43 | |
| O1:KUT tdh+ trh+ | DIE12 052499 | A. DePaola, U.S. FDA, Dauphin Island, AL | +d | 7 | |
| V. vulnificus | VV1003 | M. L. Tamplin, University of Florida, Gainesville, FLc | +d | 33 |
The toxRS operon is a global transcriptional activator that senses changes in the environment and coordinately regulates the expression of multiple genes, including those encoding multiple virulence factors and outer membrane proteins and those involved in both the acid and bile stress responses. The rpoS gene encodes a stress response sigma factor. The tdh gene is a thermostable direct hemolysin gene, and trh is a tdh-related hemolysin gene. KUT, K untypeable.
Str, streptomycin-resistant strain.
M. L. Tamplin is currently at the University of Tasmania, Hobart, Australia.
Selected for streptomycin resistance.
Two sources of algae were used in these studies. The first was a live alga, Tetraselmis chui, produced and provided by B. J. Landau at Rutgers University, Multispecies Aquaculture Demonstration Facility, Cape May, NJ. It served as a food source in oyster trials. These cultures were transported by overnight FedEx to the laboratory and used the same day they were received. Algal counts and viability were assessed microscopically using a hemocytometer. The second alga was commercially produced as a food source for shellfish (Instant Algae Shellfish Diet 1800; Reed Mariculture, Campbell, CA) and consisted of four inactivated algae (Isochrysis, Pavlova, Thalassiosira weissflogii, and Tetraselmis). They were used in studies where daily applications of algae were required.
Oyster and seawater sources.
Seawater for studies on Vibrio uptake by oysters was obtained from the University of Delaware Marine Laboratory in Lewes, DE. The samples of seawater were collected from incoming tides, which gave low-sediment seawater at approximately 30 ppt salinity. Surface water was also collected in sterile 20-liter bottles from a dock at the Cape May-Lewes Ferry Terminal in Lewes, DE, which was also around 30 ppt salinity. Screening for predatory bacteria against V. parahaemolyticus was performed on seawater samples provided by the following: William Berkhardt, U.S. Food and Drug Administration (FDA), Dauphin Island, AL; Ronald Lau, Kona Coast Shellfish, Kailua-Kona, Hawaii; Chris Langdon and Jonathan Sun, Hatfield Marine Science Center, University of Oregon, Newport, OR; and Sue Cudd, Whiskey Creek Shellfish Hatchery, Tillamook, OR. Eastern oysters (Crassostrea virginica) were obtained from John Ewart, University of Delaware Marine Laboratory, Lewes, DE, where he maintained them in a dedicated shellfish-holding facility containing large tanks of shellfish with flowthrough, natural seawater from the Broadkill River at approximately 30 ppt salinity and at ambient temperature. The outer shells were scrubbed clean to remove mud, sea-life, etc. Oysters were collected as needed and immediately transported to the laboratory in insulated coolers at ambient temperatures. They were placed in natural seawater within 2 h of collection.
Seawater was also collected and compared for Vibrio predators from four distinct Delaware sites as follows. Site 1 is the Cape May-Lewes Ferry Terminal in Lewes, DE (38°46′57.85″N; 75°07′04.73″W), which is the southernmost site directly along a rocky shoreline along the Atlantic Coast. Three sites (sites 2 to 4) are riverine sites. Site 2 is the University of Delaware oyster maintenance facility on the Broadkill River in Lewes (38°47′26.37″N; 75°09′51.36W), which is 0.6 km upstream from Roosevelt Inlet on the Delaware Bay; the water there generally has high salinity. Site 3 is a boat landing on Oyster Rocks Road, Milton, DE (38°48′08.01″N; 75°12′11.57″W), which is also on the Broadkill River 4.5 km upriver from the Delaware Bay. Site 4 is Scotton Landing on the Saint Jones River, Frederica, DE (39°05′05.94″N; 75°27′39.99″W), which is the northernmost site and 6.2 km from the Delaware Bay. The water at Scotton Landing (site 4) on the Saint Jones River generally has low salinity, while the landing at Oyster Rocks Road on the Broadkill River (site 3) is a location which has extensive marshlands immediately surrounding the site. The sites are within 10 km of each other, with the exception of Scotton Landing, which is about 45 km northwest of the southernmost site.
Development of a pour plate method for V. parahaemolyticus and V. vulnificus quantification.
A pour plate method was developed and evaluated for quantifying wild-type V. parahaemolyticus O3:K6 (RIMD2210633) and ΔtoxRS and ΔrpoS mutants, V. parahaemolyticus O1:KUT, and V. vulnificus strain VV1003 in seawater, shellfish, and pure culture. Streptomycin-resistant strains were selected for each Vibrio on LB–3% NaCl plates containing 25 μg streptomycin (Sigma Chemical Co., St. Louis, MO) per ml of agar and quantified in stock cultures, seawater, and oysters, by a pour plate assay. For the pour plate assay, we boiled, dissolved, and sterilized LB–3% NaCl agar, and while the agar was hot, we aseptically added 20-ml portions of agar to 30-ml sterile, screw-cap tubes. The tubes were maintained in a 48°C water bath until use. Streptomycin was diluted in autoclaved water and sterile filtered through a 0.22-μm-pore-size syringe filter to give a stock of 200 mg/ml, which was maintained at 4°C until use. Pour plates were prepared by adding 40 μl of streptomycin stock (determined to be the optimum amount) to the warm LB–3% NaCl tubes of agar followed immediately by the addition of 1 ml of test sample (seawater, oyster homogenate, stock Vibrio culture [to determine titers], or serial 10-fold dilutions in 0.1% peptone buffer containing 3% NaCl). Each tube was immediately inverted 3 times to mix and then poured into a 100-mm petri dish to solidify. Dishes were incubated at 37°C for 24 h ± 2 h. Vibrio colonies were enumerated under the magnification of a Quebec dark-field colony counter (Leica, Buffalo, NY).
To determine whether short-term exposure of wild-type V. parahaemolyticus O3:K6 to the 48°C agar used in the pour plate assay caused a possible reduction in Vibrio counts, the heat sensitivity of V. parahaemolyticus O3:K6 was evaluated. We compared a conventional spread plate assay (performed without heating the cultures) with a pour plate assay (performed by adding 1 ml of culture to 48°C medium) both performed with LB–3% NaCl agar containing 25 μg streptomycin per ml. V. parahaemolyticus O3:K6 was grown in LB–3% NaCl broth at 37°C to an optical density at 600 nm (OD600) of 0.060 ± 0.001 and diluted to 10−6 in 0.1% peptone buffer containing 3% NaCl, and 1-ml portions of each dilution were plated in triplicate by the pour plate method, while 0.1 ml was used for the spread plate method. The plates were incubated for 24 h at 37°C, the colonies were enumerated, and counts (numbers of colonies per milliliter of culture) were calculated. Three independent assays were performed in triplicate (n = 9).
Oyster challenge study.
Experiments were performed during the spring, summer, and fall over a 2-year period using freshly collected oysters to determine whether the wild-type strain and ΔtoxRS or ΔrpoS mutants of V. parahaemolyticus would persist in seawater or be bioconcentrated in oysters. For each experiment, four 40-liter aquaria were thoroughly cleaned and placed in a specially designed biocontainment hood. Twenty liters of natural seawater ranging from 21 to 28°C and from 25 to 30 ppt salinity was added to each aquarium along with 15 oysters obtained from seawater of approximately the same temperature and salinity. The oysters were placed on plastic, open-mesh platforms about 10 cm off the bottom of the tanks so their feces would drop freely from the shellfish. Seawater was oxygenated in each tank using typical aquarium air stones, and new air stones were used for each experiment. The oysters were allowed to acclimate overnight and to consume any algae or other food that was present in the natural seawater. All experiments were performed at room temperature (21 to 25°C).
In preparation for tank inoculations with the vibrios, LB–3% NaCl agar plates were streaked with cultures of each strain and incubated overnight. Two colonies from each strain were selected, transferred to 20 ml of LB–3% NaCl broth, and grown at 37°C and 250 rpm to an OD600 of 0.06. Aquaria containing acclimated oysters were inoculated with 0.5 ml of the appropriate Vibrio strain (approximately 5.0 × 107 bacteria/tank or 2.5 × 103 bacteria/ml of seawater), while a negative-control aquarium with oysters remained uninoculated. Live algae, T. chui, were also added to each tank to a final count of approximately 5 × 103/ml of seawater to stimulate oyster pumping. Pour plate assays were performed with 1 ml of undiluted seawater collected at 0 h and with seawater serially diluted 1:10 and higher in 0.1% peptone–3% NaCl buffer. Three independent experiments were performed with three water samples taken for each experiment, and each sample was assayed in triplicate at various dilutions (n = 27 for each dilution). Seawater from each aquarium was also collected at 24, 48, and 72 h, and similar pour plate assays were performed (n = 27 for the countable dilutions at each time interval). At 24, 48, and 72 h, three oysters from each tank were shucked, diluted 1:5 with 0.1% peptone–3% NaCl buffer, homogenized, and serially diluted in the same buffer. For each experiment, homogenate was sampled 3 times and diluted to as high as 10−6, and 1 ml of each dilution was assayed by the pour plate technique in triplicate. Three independent experiments were performed (n = 9). The above procedure was repeated with V. parahaemolyticus strain O1:KUT and with V. vulnificus strain VV1003.
Growth of vibrios in autoclaved seawater.
Vibrios did not persist in natural seawater, so seawater was autoclaved for 30 min at 121°C to determine whether autoclaving removed the Vibrio inhibitors. The screening was intended to determine whether the inhibitor was heat stable, to suggest the possible presence of a heat-stable toxin or chemical contaminant, or heat labile, which would suggest that the inhibitor was of microbial origin. Autoclaved seawater samples (100 ml each) were placed in 250-ml flasks, spiked with 6.25 μl of a culture of wild-type V. parahaemolyticus O3:K6 or the KUT strain or with V. vulnificus at an OD600 of 0.06 to give approximately 2.5 × 105 vibrios/ml of seawater to which air stones and sterile hoses were added. The flasks were incubated at room temperature with constant aeration. Water was collected at 0 h and at approximately 24-h intervals for 72 h and serially diluted water samples were tested for vibrios by the pour plate assay.
Plaque assay for VPB.
A double-agar-layer plaque assay procedure was employed to screen for the potential presence of Vibrio predatory bacteria (VPB) in seawater samples and enrichments. The procedure used polypeptone peptone (Pp) plus Bacto agar (both from Becton, Dickinson and Co.) to give Pp 20 medium, as previously reported for the detection of BALOs (45, 46); however, the volume of seawater added to the medium was reduced to prevent excessive surface liquid and softness of the agar plates. The modified formulation for Pp 20 agar was as follows: 1 g/liter Pp plus Bacto agar at the rate of 15 g/liter for the bottom agar layer or 7.5 g/liter for the top agar layer, dissolved in seawater that had been autoclaved and filtered through a 500-ml, 0.22-μm filter unit (Nalgene Nunc International Corp., Rochester, NY). The seawater was obtained from the Delaware coast and was ∼30 ppt salinity. Host vibrios were grown in LB–3% NaCl broth until they reached an OD600 of ∼0.20 (∼1.8 × 108 CFU/ml). For each assay, 25 ml of bottom-layer Pp 20 agar was dispensed into a 100-mm by 15-mm petri dish and allowed to harden. Top agar was pipetted into sterile glass tubes (7.5 ml/tube) while the agar was hot and allowed to cool to 48°C in a water bath. For the analysis of VPB in environmental seawater, 500 ml of test seawater was first filtered through a 0.45-μm, 500-ml filter to remove particulates and most bacteria. The plaque assay was conducted by combining 1 ml of host V. parahaemolyticus culture (at an OD600 of 0.20) and 7.5 ml of test (filtered) seawater to 7.5 ml of molten (48°C) Pp 20 agar in tubes. The tubes were inverted 3 times to mix and poured on top of the existing bottom layer. The plates were incubated at 22 to 26°C for up to 7 days and observed daily for clear plaques within a lawn of Vibrio host cells. Selected plaques were picked with a sterile, cotton-plugged, Pasteur pipet fitted with an eyedropper bulb to 100 μl of sterile seawater and stored at room temperature in the dark. Further plaque assays of plaque-purified isolates were performed using 1 μl of seawater containing the previously picked material (or dilutions thereof), 7.5 ml of sterile seawater, 7.5 ml of molten Pp 20 agar, and 1 ml of V. parahaemolyticus host cells in the top layer. Plaque purifications were performed from isolated plaques at least 4 times before the plaques were examined by electron microscopy.
Comparison of Vibrio and VPB levels.
Once it was determined that VPB were involved in Vibrio reductions in seawater and oysters, experiments on the uptake and persistence of V. parahaemolyticus O3:K6 were conducted. In these experiments, 20 liters of natural seawater containing 15 oysters was inoculated with 2.5 × 103 vibrios/ml for subsequent analysis of the seawater for both V. parahaemolyticus O3:K6 and VPB levels at 0, 24, 48, and 72 h. Oysters were also tested daily for vibrios, but not for VPB, since methods have not been developed for the quantitative detection of VPB in oyster meats. Immediately after drawing 0-h water samples for Vibrio and VPB analyses and 0-h oyster samples for Vibrio analysis, oysters remaining in the tanks were fed a ration of Instant Algae Shellfish Diet 1800 (Reed Mariculture, Campbell, CA) consisting of four inactivated algae (Isochrysis, Pavlova, Thalassiosira weissflogii, and Tetraselmis) at the rate of 1 × 106 algae/ml of seawater. Algae were added to ensure that the oysters were actively feeding. Feeding was repeated at the same rate at 24 h but was reduced to 5 × 105 algae/ml of seawater at 48 h, since a portion of the oysters had been removed for daily testing. Three water samples were collected for analysis at each time point. Samples were diluted in 0.1% peptone buffer containing 3% NaCl. Seawater and diluted water samples were each tested for V. parahaemolyticus O3:K6 and VPB in triplicate, and each trial was performed twice (n = 18). Three oysters were also collected at each time point, shucked, combined, homogenized, and serially diluted in 0.1% peptone buffer, and each dilution was analyzed for V. parahaemolyticus O3:K6 in triplicate daily for 72 h. This trial was performed twice (n = 6). For each trial, a tank with seawater and 15 oysters, but without added V. parahaemolyticus, was used as endogenous Vibrio (negative) controls and were analyzed in the same manner as the tanks containing Vibrio-challenged oysters and seawater.
Electron microscopy.
Scanning electron microscopy was performed on stock cultures of Bdellovibrio bacteriovorus and Bacteriovorax stolpii in E. coli host cells and on enrichments from plaque-purified seawater isolates grown in V. parahaemolyticus O3:K6. V. parahaemolyticus and E. coli were propagated as described above. Vibrios and enrichments for VPB were often diluted in autoclaved and 0.22-μm-filtered seawater 1:100 and 1:10, respectively, just prior to slide preparation. In addition, VPB enrichments were occasionally spiked with small amounts (10 to 20% total volume) of fresh host cells to examine the initial interactions between the organisms. Preparative techniques included the following: inoculation of 200 μl of sample onto a coverslip; adherence of bacteria to the coverslip at room temperature for 30 min; fixation of isolates on slides by the addition of 20 μl of 25% glutaraldehyde (Electron Microscopy Sciences, Fort Washington, PA) to the 200-μl samples for 1 h; rinsing with 0.1 M imidazole buffer (pH 7.0) for 30 min; and dehydration in increasing ethanol rinses (1 time at 20% and 2 times at 30, 50, 80, 90, and 100% for 30 min each). The preparations were subjected to critical point drying in a Denton vacuum critical point dryer (Denton, Cherry Hill, NJ) according to the manufacturer's instructions and shadowed with gold 3 times for 30 s each time on an Edwards Scancoat Six sputter coater (West Sussex, United Kingdom) in an argon atmosphere. Images were viewed under a Quanta200FEG scanning electron microscope (FEI Co., Hillsboro, OR).
Screening for VPB from Atlantic, Gulf, and Pacific seawater.
Seawater samples obtained from Delaware, Alabama, Oregon, and Hawaii were evaluated for the presence of VPB. Plaque assays for VPB were performed using V. parahaemolyticus O3:K6 as host cells. The samples of seawater were filtered through 0.45-μm-pore-size filters, and 7.5-ml portions were analyzed by the VPB plaque assay described above. Although the filtration was expected to remove most of the bacterial contaminants and some of the VPB, a portion of VPB would be expected to pass through 0.45-μm-pore-size filters. Filtration was required to prevent overgrowth of the plaque assay plates by non-Vibrio spreaders. Thus, this assay will underestimate the true levels of VPB in the seawater. Delaware seawater samples were also examined for fluctuations in filterable VPB counts monthly from four sites from October to March, and the samples were assayed by plaque assay in triplicate.
Phage plaque assay.
Presumptive VPB isolates were screened to determine whether any were bacteriophages. The phage plaque assay was performed using a double-agar procedure. The bottom agar consisted of the following (per 100 ml): 1.0 g tryptone (Fisher Scientific, Park Lawn, NJ), 2 g NaCl, 0.10 g dextrose (Fisher Scientific), 1.5 g of Sigma type II agarose (Sigma-Aldrich, St. Louis, MO), and 100 ml distilled H2O (dH2O). Fifteen milliliters of autoclaved bottom agar was added per 100-mm-diameter petri dish. The top agar consisted of the following (per 100 ml): 1 g tryptone, 2 g NaCl, 0.1 g dextrose, 0.5 g yeast extract (Fisher Scientific), 0.037 g CaCl2 · 2H2O (Sigma-Aldrich), 0.05 g MgCl2 · 6H2O (Fisher Scientific), 0.6 g Sigma type II agarose, and 100 ml of dH2O. Ten milliliters of freshly autoclaved, hot agar was aseptically dispensed into sterile 15-mm screw-cap glass culture tubes and maintained in a water bath at 48°C. LB–3% NaCl broth was inoculated with a colony of V. parahaemolyticus O3:K6 and grown to an OD600 of 0.6. Then, 250 μl of the culture was added to the tube of warm top agar along with 1 ml of the cultured isolate. Each tube was manually mixed by inversion 3 times and poured over the bottom agar layer. A known V. parahaemolyticus O3:K6 phage isolated by this laboratory was used as a positive control. After solidification, the plates were covered, inverted, stored at room temperature, and observed for plaques for up to 5 days.
Statistical analyses.
Vibrio uptake and depletion data for oysters and seawater was evaluated by analysis of variance (ANOVA) using Proc Mixed of the SAS software system (SAS Institute Inc., Cary, NC). Mean separations were performed using the least significant difference (LSD) and Dunnett test methods to determine the significance of the comparisons. These analyses were performed using the log10 of the raw counts, with a randomly generated count between 0 and the limit of detection for zero counts. The Dunnett test was performed to compare relationships between the wild-type V. parahaemolyticus O3:K6 and ΔtoxRS counts and between the wild-type and ΔrpoS counts at each time interval. Paired Student's t tests were also used to determine whether differences in uptake in oysters or in persistence in oysters and seawater were significant between the Vibrio species. P values of <0.05 were considered statistically significant.
RESULTS
Enumeration of vibrios.
A simple, rapid, and reliable pour plate assay was developed to quantify streptomycin-resistant vibrios as described in Materials and Methods. Streptomycin-resistant Vibrio counts as high as 300 CFU could easily be enumerated on each 100-mm-diameter petri dish, due to the restricted size of the predominantly subsurface colonies (approximately 0.5- by 1-mm elliptical colonies). The counts were comparable in plates containing no antibiotic and plates containing as high as 40 μl of streptomycin stock in 20 ml of medium (400 μg streptomycin/ml); thus, 400 μg streptomycin/ml of medium was determined to be optimal and was used throughout this study. Streptomycin reduced background bacteria in oysters and seawater to nondetectable levels.
Short-term exposure of wild-type V. parahaemolyticus O3:K6 to the 48°C agar used in the pour plate assay was evaluated to determine whether heat negatively affected Vibrio counts. We compared a conventional spread plate assay (performed without heating the cultures) with the pour plate assay (performed by adding culture to 48°C medium) and obtained significantly higher counts/ml of culture (5-fold higher; [P < 0.0001]) with the pour plate method. We conclude that our pour plate assay is superior to spread plating and that the brief exposure to tempered agar did not negatively affect counts.
Comparison of survival of V. parahaemolyticus O3:K6 and ΔtoxRS and ΔrpoS mutants in natural seawater and oysters.
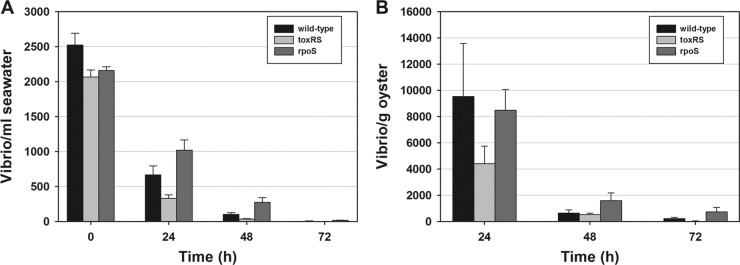
The introduction of approximately 5 × 107 wild-type V. parahaemolyticus O3:K6 and the corresponding toxRS or rpoS deletion mutant into 20 liters of natural seawater containing 15 oysters and live algae led to a rapid and significant (P ≤ 0.05) daily decrease in V. parahaemolyticus counts in the seawater over 72 h (Fig. 1A). This decrease was originally thought to be from Vibrio uptake by the oysters, but oyster analyses showed low levels of viable vibrios within the tissues after uptake for 24 h followed by rapid and significant (P ≤ 0.01) count reductions after 24 h (Fig. 1B). Note that 0-h counts in oyster homogenates were always nondetectable (<5 CFU/ml [data not shown]). In oysters, the numbers of ΔtoxRS mutants were lower than the numbers of the wild type and ΔrpoS mutant at 24 h (Fig. 1B); however, differences were not significant (P > 0.05). In natural seawater, 73.6%, 84.0%, and 52.8% of the wild-type, ΔtoxRS, and ΔrpoS strains were lost within 24 h, respectively. At 48 h, count reductions were ≥96%, except for the ΔrpoS mutant, which was reduced by 87.5%. By 72 h, <1% of the three Vibrio strains remained in the seawater. Oysters accumulated vibrios over 24 h, but the counts decreased significantly over the next 24 h (P < 0.05) with <20% of the ΔtoxRS and ΔrpoS mutants remaining in the oysters, and only 5.9% of the wild type remained. By 72 h, counts had decreased by ≥97.8% from the 24-h highs for the wild type and ΔtoxRS mutant, while ΔrpoS levels diminished by 91.5% for that same period. Comparison of total V. parahaemolyticus levels that accumulated in the oysters after 24 h showed that only 2.9% of the wild type, 1.5% of the ΔtoxRS mutant, and 2.8% of the ΔrpoS mutant present in the water were detected in the oysters, even though the oysters were observed to be actively pumping during each experiment, as evidenced by the production of feces and pseudofeces each day, shell gaping, and clearing of the algae from the water.
Fig 1.
Levels of viable vibrios in seawater and shellfish and their persistence over time. (A and B) Counts of wild-type Vibrio parahaemolyticus O3:K6, toxRS deletion mutants, and rpoS deletion mutants in seawater (A) or in oysters maintained in tanks of seawater (B). The error bars in panel A indicate the standard errors of the means (SEM) of three independent experiments, each performed three times in triplicate (n = 27), while the error bars in panel B represent the SEM of three independent experiments each performed in triplicate (n = 9).
We calculated the approximate total levels of V. parahaemolyticus in the tank system (oysters plus seawater) at 24 h and determined that only 29.3% of the initial inoculum of the wild-type strain, 17.4% of the ΔtoxRS mutant, and 50.1% of the ΔrpoS mutant remained in each system at 24 h. These values are slightly greater than the percentages remaining in the water alone after 24 h (26.4, 16.0, and 47.2%, respectively). These results indicate a failure of V. parahaemolyticus O3:K6 and the deletion mutants to persist in seawater or within shellfish. For both seawater and oysters, the Dunnett test showed that there was no significant difference in Vibrio counts between the ΔtoxRS mutant and the wild type or the ΔrpoS mutant and the wild type at 24, 48, or 72 h. Overall, we concluded that the toxRS and rpoS genes did not convey a selective advantage for the uptake or persistence of V. parahaemolyticus O3:K6 in oysters or their persistence in natural seawater.
Vibrio parahaemolyticus O1:KUT and V. vulnificus in seawater and oysters.
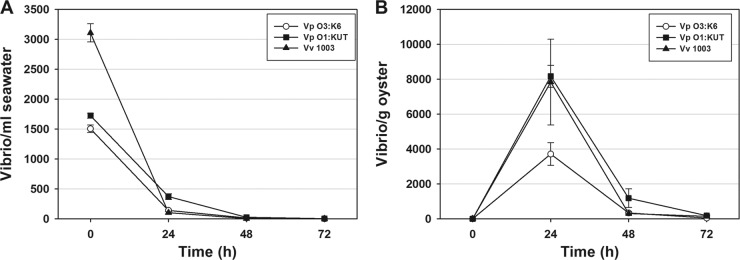
Since V. parahaemolyticus O3:K6 and the deletion mutants failed to persist in natural seawater or oysters, we evaluated whether this failure to thrive was unique to this species and serotype. We compared the levels of V. parahaemolyticus O3:K6, V. parahaemolyticus O1:KUT, and V. vulnificus in seawater and their uptake and persistence in oysters. Comparable results were obtained for all three pathogens in seawater with all vibrios decreasing significantly (P < 0.05) by 24 h (Fig. 2A). Oysters bioaccumulated limited numbers of all three vibrios by 24 h; however, counts significantly decreased (P < 0.05) for the three pathogens by 72 h (Fig. 2B). These results showed the inability of these pathogens to persist in natural seawater or oysters.
Fig 2.
Comparison of the levels of V. parahaemolyticus (Vp) O3:K6, V. parahaemolyticus (Vp) O1:KUT, and V. vulnificus VV1003 (Vv 1003) in seawater (A) and oysters (B) over 72 h. The error bars in panel A indicate the SEM of three independent experiments with three subsamplings per time point with assays performed in triplicate (n = 27), while the error bars in panel B indicate the SEM of three independent experiments each performed in triplicate (n = 9).
Seawater studies.
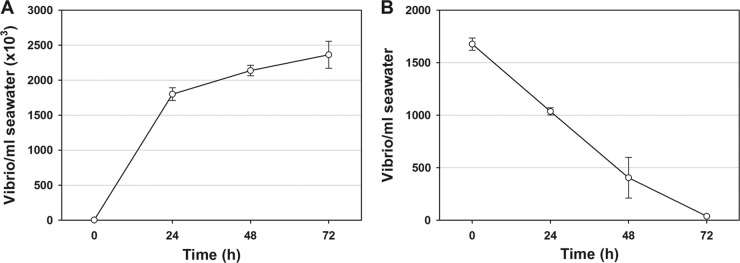
To determine whether the reductions in Vibrio levels from the levels in natural seawater were likely due to the uptake of the bacteria by the oysters through their natural filtering process, V. parahaemolyticus O3:K6 was added to natural seawater (without oysters or added algae), and Vibrio levels in natural water were monitored daily for 72 h. Results indicate a steady decline of V. parahaemolyticus from starting levels of approximately 1.5 × 103 to near negligible levels by 72 h (Fig. 3). Since no oysters were included in these experiments, some factor in the seawater was responsible for the Vibrio reductions.
Fig 3.
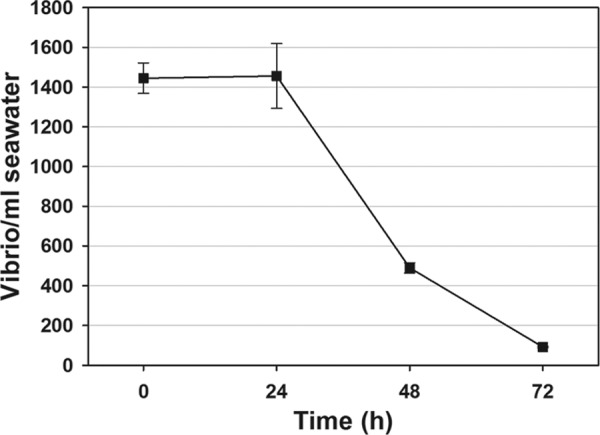
Reduction of V. parahaemolyticus O3:K6 in natural seawater without oysters. The error bars represent SEM of three independent experiments performed in triplicate (n = 9).
Autoclaved seawater.
To begin to identify the cause of Vibrio decline in seawater (with or without oysters), we examined whether the inhibitory factor was heat labile. In autoclaved seawater, the starting levels of V. parahaemolyticus O3:K6 were 2.0 × 103 CFU/ml and increased to 2.4 × 106 CFU/ml over 72 h for a 1,000-fold increase (Fig. 4A). A similar starting level of V. parahaemolyticus in natural seawater (1.7 × 103 CFU/ml) led to a reduction to only 36 CFU/ml by 72 h for a 47-fold reduction in counts (Fig. 4B). These results suggested that Vibrio inhibition was caused by the presence of a heat-labile inhibitor, possibly of microbial origin; therefore, we sought to identify the possible presence of BALOs or other predatory bacteria.
Fig 4.
Comparison of streptomycin-resistant V. parahaemolyticus O3:K6 counts in autoclaved seawater (A) and natural seawater (B) over 72 h. Note the different scales of the y axes and that the mean starting levels of vibrios in panels A and B were approximately the same (1.5 × 103 vibrios/ml). The error bars represent SEM of three samples tested in triplicate (n = 9).
Plaque assay for VPB.
A plaque assay was devised to quantify BALOs and other predatory organisms on V. parahaemolyticus O3:K6 host cells as described in Materials and Methods. Plaque-like foci formed on lawns of V. parahaemolyticus O3:K6 host cells. Some plaques appeared as large, yellowish clearings that increased to >2 cm in diameter within 5 days. Other isolates produced clear plaques of widely different sizes ranging from about 2 mm to >2 cm after 5 days. Isolates were picked and purified through at least 4 rounds of plaque purification, and selected isolates were visualized by scanning electron microscopy. Plaque assays for bacteriophages were also performed on the isolates, but only the positive controls produced plaques.
Comparison between vibrios and their predators in seawater and oysters.
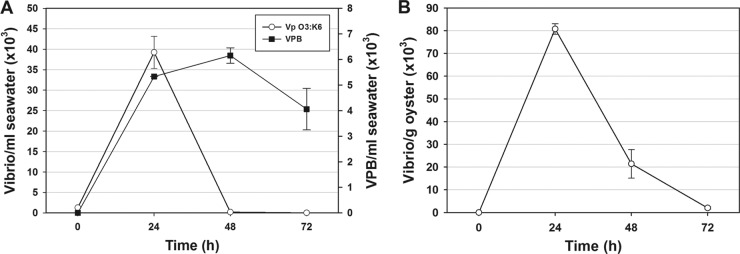
Tanks containing fresh, natural seawater and 15 oysters from the Delaware coast were inoculated with V. parahaemolyticus O3:K6, and both V. parahaemolyticus and VPB were quantified daily for 72 h after V. parahaemolyticus inoculation. Results showed the presence of nearly nondetectable levels of naturally occurring VPB in seawater at 0 h, an increase to 6.2 × 103 PFU/ml at 48 h, and some tapering off to 4.1 × 103 by 72 h (Fig. 5A). In contrast, the mean Vibrio level added to the seawater at 0 h was 1.3 × 103 CFU/ml; however, the levels increased to 3.9 × 104 CFU/ml by 24 h and then decreased to near negligible levels by 48 h as the levels of VPB increased (Fig. 5A). The levels of V. parahaemolyticus in the oysters were also monitored over 72 h (Fig. 5B) and followed the same trend as the Vibrio levels in seawater, which would suggest the inactivation of oyster-associated vibrios by VPB. The VPB levels in oysters could not be directly quantified, because methods have not been developed.
Fig 5.
Comparison of mean counts of V. parahaemolyticus (Vp) O3:K6 and Vibrio predatory bacteria (VPB) (■) over time. (A) Vibrio and VPB counts in natural seawater and (B) Vibrio counts in oysters. Two experiments were performed in triplicate (n = 9). The error bars represent SEM.
Vibrio persistence in the presence of live or inactivated algae.
Comparisons were made to determine whether V. parahaemolyticus O3:K6 counts in natural seawater varied depending on the presence of live algae (Fig. 2A) or the presence of a commercial (inactivated) alga mix (Fig. 5A). Algae had been added to stimulate oyster pumping in both experiments. With a commercial algal diet administered daily at the rate of ≥5 × 105 algae/ml of seawater, V. parahaemolyticus O3:K6 increased in natural seawater over 24 h, followed by reductions to negligible levels by 72 h (Fig. 5A). In contrast, no increases over the initial levels were observed in earlier studies which were performed with live algae added at the rate of 5 × 103 algae/ml of seawater at the beginning of the study (Fig. 2A). The initial elevation of V. parahaemolyticus levels in the inactivated alga study shown in Fig. 5A may have been associated with the following: (i) the presence of additional nutrients to support V. parahaemolyticus growth in seawater; (ii) restriction in the motility of the VPB due to the additional algae, thus making it more difficult for VPB to find its prey; or (iii) lower starting levels of VPB in this natural seawater. In oysters that were fed the commercial algal diet, V. parahaemolyticus O3:K6 levels rose over the first 24 h but decreased to negligible levels by 72 h (Fig. 5B), which is consistent with our findings where oysters were fed live algae (Fig. 2B). Thus, V. parahaemolyticus O3:K6 counts in oysters diminished to near negligible levels over the 72-h time frame in the presence of exogenously added live or inactivated algae.
Electron microscopy.
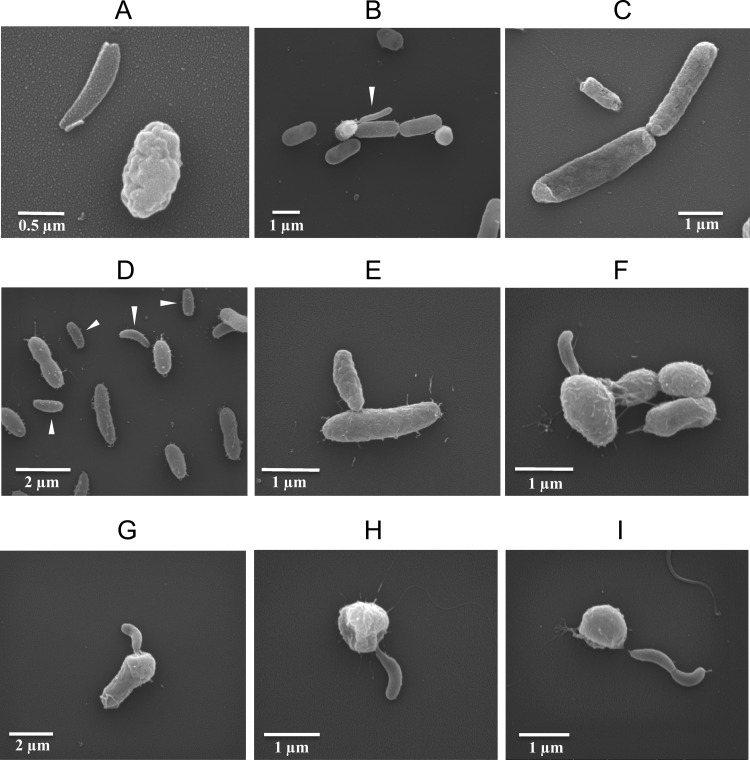
Scanning electron microscopy was performed on ATCC reference strains of Bdellovibrio bacteriovorus (ATCC 15143) and Bacteriovorax stolpii (ATCC 27052) in E. coli host cells and on plaque-purified isolates originally obtained from cultures of natural but 0.45-μm-filtered seawater in V. parahaemolyticus O3:K6 host cells. Seawater isolates displayed a variety of morphologies consistent with Bd. bacteriovorus, Ba. stolpii, and Micavibrio aeruginosavorus. The Bd. bacteriovorus reference strain is shown in Fig. 6A to C, and associated seawater isolates are shown in Fig. 6D to I. The immature form of Bd. bacteriovorus is relatively smooth and slender, as depicted in Fig. 6A (left) and B (white arrowhead). In contrast, a mature, attack phase form is generally larger/broader and more irregularly shaped, as shown in Fig. 6A (right) and in Fig. 6C, where one attack phase cell is seen along with two larger E. coli hosts as they divide. It should be noted that E. coli and V. parahaemolyticus exhibited some elongated cells when grown in medium containing seawater. Representative isolates that resemble BALOs are shown in Fig. 6D to I. Short, host-dependent forms consisting of suspected attack phase BALOs are indicated by white arrowheads (Fig. 6D). Attack phase Bd. bacteriovorus bacteria are reportedly 0.2 to 0.4 μm in diameter and 0.5 to 1.4 μm in length with a single polar flagellum (45), which is consistent with the sizes observed in Fig. 6. Flagella were difficult to see in most micrographs. V. parahaemolyticus O3:K6 was used to propagate the isolates seen in Fig. 6D to I and range between 0.5 and 0.8 μm in diameter and 1.4 and 2.4 μm in length (37), as expected.
Fig 6.
Representative scanning electron micrographs of plaque-purified predatory bacteria. (A to C) Bdellovibrio bacteriovorus (ATCC 15143) propagated in E. coli host cells. (A) An immature Bd. bacteriovorus (left) and an attack phase Bd. bacteriovorus (right). (B) Bd. bacteriovorus (white arrowhead) emerging from a host E. coli. (C) Attack phase Bd. bacteriovorus (flagellum visible) approaching a somewhat elongated E. coli. (D to I) Plaque-purified isolates cultured from 0.45-μm-filtered seawater in V. parahaemolyticus O3:K6. (D) Bdellovibrio and like organisms (BALOs) (white arrowheads) amid a background of larger V. parahaemolyticus O3:K6 host cells. (E) Attack phase BALO entering a V. parahaemolyticus host cell. (F to G) Immature BALOs exiting swollen V. parahaemolyticus host cells. (H) Immature BALO exiting spent V. parahaemolyticus. (I) Conjoined BALOs exiting a spent V. parahaemolyticus.
A small, attack phase BALO appears to be attached to a host cell in Fig. 6E, pending entry into the host, while Fig. 6F to I display apparent immature BALOs exiting the cells. Note the rounded appearance of the V. parahaemolyticus hosts as the immature BALOs exit. The fattened diameter of the V. parahaemolyticus in Fig. 6F and G suggest the presence of an enlarged bdelloplast containing multiple immature cells awaiting release from the host, although the internal structure of host cells could not be examined by scanning electron microscopy. Figure 6H shows an apparent immature BALO exiting a “spent” (used up) V. parahaemolyticus host cell. Many of the parasitized vibrios took on this rounded appearance. A novel, double, immature BALO appears to have recently been released from a rounded and spent V. parahaemolyticus in Fig. 6I. Other plaque-purified isolates showed no signs of internalization into the host but still parasitized their hosts to produce plaques, consistent with the extracellular parasitism caused by Micavibrio aeruginosavorus-like predators. Some of our VPB isolates may in fact be M. aeruginosavorus, which resembles attack phase BALOs (21).
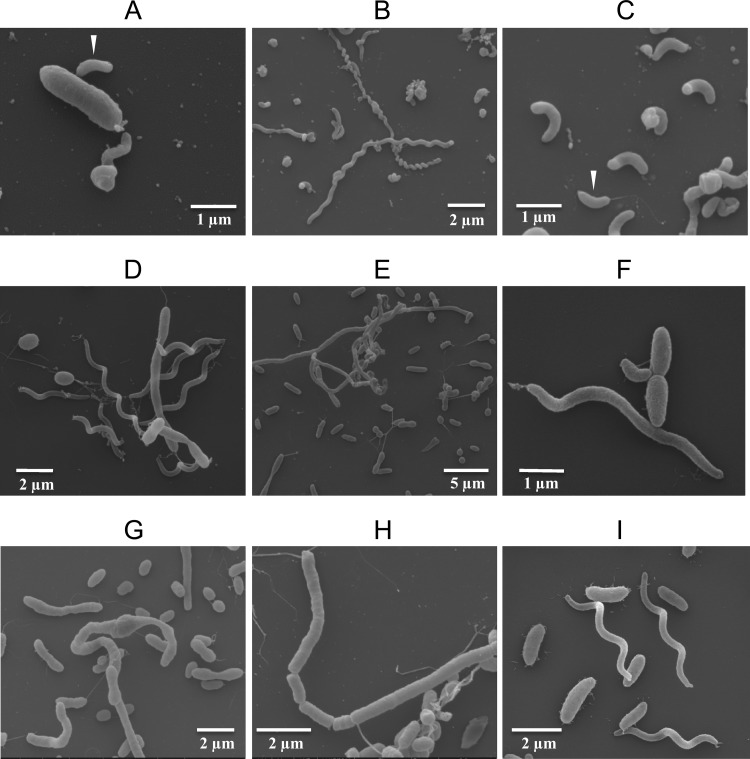
The attack phase Bacteriovorax stolpii is reportedly the same size as Bdellovibrio bacteriovorus and also has a single polar flagellum (43). A Ba. stolpii stock culture (ATCC 27052) shows at least one host-dependent, attack phase bacterium entering an E. coli cell (Fig. 7A, white arrowhead); long, host-independent, spiral forms (Fig. 7B); and shorter, curly forms with bulbous ends (Fig. 7C). Note the attack phase form with a single polar flagellum indicated by the white arrowhead (Fig. 7C). Among the plaque-purified seawater isolates were forms with these same morphological characteristics (Fig. 7D to I). Some isolates had filamentous forms in excess of 100 μm in length (not shown). Seawater isolates shown in Fig. 7D to I had been filtered through 0.45-μm-pore-size filters before enrichment and plaque assay. The presence of elongated and/or spiral forms in filtered seawater signifies de novo synthesis of these forms. Many plaque-purified VPB isolates did not conform morphologically to known BALOs but were clearly parasitic toward V. parahaemolyticus. They are the subject of ongoing investigations.
Fig 7.
(A to C) Scanning electron micrographs of stock culture of Bacteriovorax stolpii (ATCC 27052) showing their small, attack phase form (white arrowhead) infecting a host E. coli (A), host-independent spiral filaments (B), and short forms with bulbous ends (C). A typical, flagellated, attack phase form (white arrowhead) is also s in panel C. (D to I) Isolates of plaque-purified Ba. stolpii-like organisms cultured in V. parahaemolyticus from 0.45-μm-filtered seawater.
VPB screening of Atlantic, Gulf, and Pacific seawater.
Samples of seawater from the Atlantic Coast of Delaware, the Gulf Coast at Dauphin Island in Alabama, two sites along the Pacific Coast in Oregon, and from Kailua-Kona, Hawaii, were assayed for Vibrio predators. Testing of 7.5 ml of nonenriched, 0.45-μm-filtered seawater samples revealed the presence of plaques against V. parahaemolyticus from water from Delaware, Alabama, and Hawaii; however, no plaques were isolated from seawater obtained from two shellfish hatcheries in Oregon. To obtain some idea of the levels of VPB in Delaware seawater over time, samples were collected monthly from four sites from October to March. The water was filtered through a 0.45-μm filter and analyzed for VPB by a plaque assay. The numbers of VPB and associated seawater temperatures and salinities at the time of seawater collection are shown in Table 2. Since plaque assays are designed to test 7.5 ml of 0.45-μm-filtered seawater, results are expressed as mean PFU per 7.5 ml of seawater for tests performed in triplicate. Results show mean levels as high as 23.6 PFU/7.5 ml of seawater for one sampling site (Table 2). Variability in VPB levels may have been related to fluctuations in salinities and temperatures among the sites, although there was insufficient information to correlate salinity or temperatures with the number of bacteria (Table 2). Salinities ranged from 1.8 to 29.5 ppt, while temperatures ranged from 5.5 to 19.4°C. Plaque counts during the cooler months were generally low, with the mean counts from sites 1 to 4 being 2.5, 1.5, 5.5, and 0.2 PFU/7.5 ml of seawater, respectively. The highest levels of VPB were from site 3, which is a high-salinity riverine site with extensive marshlands on both sides of the river, suggesting that VPB may be associated with high-productivity marshes. In order to better evaluate fluctuations in VPB levels throughout the year, testing will continue at these sites. Absolute quantification for VPB was not possible in seawater screening, since natural seawater was first filtered through a 0.45-μm-pore-size filter to remove many of the bacterial contaminants which could otherwise overgrow the plaque assay plate. This filtration would be expected to allow smaller VPB, particularly BALOs in their immature or attack phase forms, and similarly sized M. aeruginosavorus to pass through the filters for analysis. Larger isolates, like the filamentous and spiral morphotypes of BALOs, cannot pass through 0.45-μm filters and, therefore, could not be enumerated or included in Table 2 data.
Table 2.
Quantification of Vibrio predatory bacteria detected in natural seawater at four locations in Delaware from October 2011 to March 2012
| Locationa | Date (day-mo-yr) | Temp (°C) | Salinity (ppt) | Mean PFUb |
|---|---|---|---|---|
| Site 1. Cape May-Lewes Ferry Terminal, Lewes, DE | 20-10-2011 | 19.4 | 24.2 | 0.7 |
| 15-11-2011 | 12.8 | 27.9 | 13 | |
| 22-12-2011 | 9.1 | 24.6 | 0 | |
| 19-1-2012 | 8.2 | 28.4 | 0.3 | |
| 12-2-2012 | 6.2 | 26.0 | 0.7 | |
| 8-3-2012 | 9.2 | 29.5 | 0 | |
| Site 2. University of Delaware Marine Lab, Lewes, DE | 20-10-2011 | 19.4 | 25.4 | 1 |
| 15-11-2011 | 12.8 | 26.5 | ND | |
| 22-12-2011 | 8.8 | 24.0 | 4.3 | |
| 19-1-2012 | 7.9 | 28.0 | 0.3 | |
| 12-2-2012 | 6.4 | 26.4 | 1.7 | |
| 8-3-2012 | 8.1 | 29.2 | 0 | |
| Site 3. Oyster Rocks Road, Milton, DE | 20-10-2011 | 19.4 | 24.0 | 1.7 |
| 15-11-2011 | 12.5 | 26.4 | 23.6 | |
| 22-12-2011 | 7.5 | 14.7 | 2 | |
| 19-1-2012 | 7.8 | 27.5 | 0.3 | |
| 12-2-2012 | 6.1 | 25.9 | 5.3 | |
| 8-3-2012 | 9.0 | 28.8 | 0 | |
| Site 4. Scotton Landing, Frederica, DE | 20-10-2011 | 18.9 | 12.6 | 0.3 |
| 15-11-2011 | 12.9 | 11.3 | 0 | |
| 22-12-2011 | 6.5 | 1.8 | 0 | |
| 19-1-2012 | 6.4 | 9.0 | 0.7 | |
| 12-2-2012 | 5.5 | 4.4 | 0.3 | |
| 8-3-2012 | 8.8 | 4.6 | 0 |
Location coordinates and descriptions are given in Materials and Methods.
The counts are the mean number of PFU of triplicate assays of 7.5 ml of 0.45-μm-filtered seawater per assay. ND, not done.
DISCUSSION
The original aim of this study was to determine whether toxRS or rpoS played a role in colonization and persistence of Vibrio parahaemolyticus O3:K6 in shellfish. We demonstrated that V. parahaemolyticus O3:K6 failed to persist in natural seawater or oysters, regardless of the presence or absence of the toxRS or rpoS gene. Studies with other Vibrio strains, such as V. parahaemolyticus O1:KUT and V. vulnificus strain VV1003, showed that these pathogens also failed to persist in oysters or seawater containing a natural marine flora. Neither toxRS nor rpoS provided a selective advantage in the uptake of vibrios by oysters or the persistence of vibrios in oysters or natural seawater. In fact, all pathogens tested (V. parahaemolyticus strains and V. vulnificus) showed declines in seawater and limited uptake in shellfish followed by rapid decline. In autoclaved seawater, the opposite effect was observed—an increase in V. parahaemolyticus numbers (Fig. 4), suggesting that biotic factors, such as competing organisms and VPB in the natural seawater, may lead to V. parahaemolyticus decline.
Plaque assays for VPB were performed on V. parahaemolyticus O3:K6 host cells and produced an assortment of plaque morphologies. Plaque-purified isolates were examined by scanning electron microscopy and revealed a wide variety of potential BALOs and other VPB morphotypes. Data from Table 2 show marked variability in VPB levels from seawater obtained from four closely spaced locations. The highest and second highest mean counts (23.6 and 13 PFU/assay, respectively) were obtained in November from sites 3 and 1, respectively, when the seawater temperatures were 12.5 and 12.8°C, respectively, and the salinities were 26.4 and 27.9 ppt, respectively. Factors beyond temperature and salinity may influence the levels of VPBs, including the nutrient levels and oxygenation of the water and the availability of prey.
A recent study detailed the disappearance of V. vulnificus from seawater and oysters along the North Carolina coast from 2007 to 2009, a period which coincided with the worst drought in North Carolina history (11). Higher than normal salinities were observed during this period. Drought abated in late 2009 after which V. vulnificus could again be detected in seawater, but not in oysters (11). From the present study, one could hypothesize that VPBs played a role in the reduction in V. vulnificus levels during that time period. High-salinity seawater has been shown to cause reductions in V. vulnificus levels in oysters (25, 26), but the extent to which VPB may have been responsible for the decline has not been evaluated. Prey bacteria are known to shape the community structure of their predators (5). Studies on the optimal salinities for growth of our VPB isolates are under way and may lead to a better understanding of the causes of Vibrio fluctuations.
Bacteriovorax bacteria have been difficult to identify in seawater during the winter months (48), although they appear to tolerate cold temperature (5°C) storage for weeks (47). With few exceptions, we generally observed low plaque counts from seawater tested during the winter months but anticipate that much higher counts will be obtained during the summer samplings. Spatial differences were also observed among waters within close proximity to each other (Table 2), which may be related to tides, salinity, the volume of freshwater runoff, or their proximity to the open ocean or marshes. We showed that low starting levels of VPB (<1 PFU/ml of seawater) were sufficient to substantially reduce V. parahaemolyticus counts in seawater (Fig. 5A), so the low levels typically found at the Delaware sites (Table 2) would likely be sufficient to account for the reductions in Vibrio levels observed in Fig. 1 and 2. Studies quantifying seasonal VPB levels are continuing.
VPB plaques were of multiple sizes, grew at different rates, and were either clear or yellow. They all produced clear foci of infection and often had sharp, clearly defined edges, characteristic of the BALOs (45). One distinguishing mark of BALOs is that they commonly form plaques over several days, while phage plaques generally develop overnight (47). The Pp 20 medium that was used for top agar in VPB plaque assays is a low-nutrient medium which favors the development of BALOs rather than phages, which prefer more nutritionally complex media (47). A phage plaque assay was performed on isolates testing positive by VPB plaque assay, but none of the isolates produced plaques, strongly indicating that the plaques were not of viral origin. In addition, plaque picks observed by electron microscopy failed to show any signs of the presence of phage. Although no phages were observed, the occasional presence of a phage in studies involving Vibrio persistence in oysters and/or seawater could not be precluded. In a recent study, Chen and Williams (6) showed that it was possible for V. vulnificus to be coinfected by both a BALO and a phage, but such an occurrence is not expected to be common.
Electron microscopy performed on plaque-purified isolates revealed a variety of morphological structures, often within the same culture suggesting different forms of the same bacterium. The complex life cycle of BALOs can account for these forms with host-dependent and host-independent replication processes. In fact, replication may be accomplished in a host cell by formation of the bdelloplast or external to host cells or in the absence of host cells by binary fission or by septation and separation of elongated, host-independent forms (9, 20). Current dogma suggests that the elongating, replicating form within the bdelloplast is enzymatically processed into individual flagellated progeny cells before they are released from the host and that further enzymatic processing releases these progeny from the host (44). An apparent exception to this is shown in Fig. 6I, which depicts two conjoined BALOs recently released from a rounded and spent V. parahaemolyticus host cell.
One of the greatest benefits of this study was that it evaluated specific target vibrios in competition with normal marine organisms to assess the effects of competing bacteria and predatory organisms on Vibrio survival in seawater and their uptake and persistence in oysters. These studies highlight dramatic differences when using natural seawater versus autoclaved seawater. In reviewing the literature on Vibrio proliferation in seawater, fish, and shellfish, many studies used autoclaved and/or artificial seawater to eliminate endogenous vibrios. This can severely bias the results. We chose to use streptomycin-resistant Vibrio cultures so that the bacteria could be quantified after being maintained in a nonsterile, more natural setting. The use of artificial systems is often coupled with the use of highly selective media which may be inhibitory to some strains of Vibrio, especially to vibrios that are stressed. We were somewhat surprised to find that no naturally present streptomycin-resistant bacteria were detected in the seawater or shellfish; controls for streptomycin-resistant background were consistently negative. This finding may vary from one location to another, depending on population densities and antibiotic usage in the general population. Our seawater samples and oysters were obtained from rural areas not impacted by large hospitals or medical complexes. The use of streptomycin in the LB–3% NaCl agar allowed the enumeration of target bacteria, but standard spread plate assays do not permit enumeration of high numbers of vibrios, since colonies on the agar surface are large and often merge with other colonies. In contrast, our pour plate assay facilitated the enumeration of high numbers of vibrios without concern for colony size, since the vast majority of colonies were subsurface and small. In addition, pour plates could be viewed at different angles to evaluate colonies that formed at different depths in the transparent agar, thus allowing hundreds of colonies per plate to be easily and accurately enumerated. Although useful in the present studies for Vibrio enumeration, pour plate assays may not be appropriate for heat-sensitive bacteria where they could give an underestimation of bacterial populations.
To the best of our knowledge, this is the first study to quantitatively demonstrate the amelioration of V. parahaemolyticus and V. vulnificus in natural seawater and oysters due to the presence of naturally occurring predatory bacteria. This study suggests a new paradigm of the role of VPB in modulating pathogenic vibrios in shellfish in their native environment. Previously it was believed that the uptake and persistence of vibrios in oysters were dependent on physical water conditions (temperature and salinity), nutrient availability, and the oyster's ability to bioconcentrate and purge microbial contaminants as they actively pumped; however, we provide evidence that VPB likely play a direct and perhaps major role in suppressing vibrios in shellfish. Here, we provide useful methods to quantify specific pathogenic vibrios and VPB in a background of natural microflora. Future research should include the following: (i) ecological studies to identify factors that affect VPB levels in the marine environment; (ii) an evaluation of the dynamics between VPB and vibrios in their native environment; and (iii) further characterization and identification of specific VPB isolates in regard to sequence, host specificity, and life cycle.
ACKNOWLEDGMENTS
We acknowledge the excellent technical assistance of Michael Watson (U.S. Department of Agriculture [USDA], Agricultural Research Service [ARS], Dover, DE); Dallas Henry and Gulnihal Ozbay (Delaware State University, Dover, DE); and Guoping Bao, Joseph Uknalis, and John Phillips (USDA, ARS, Wyndmoor, PA). We gratefully acknowledge John Ewart (University of Delaware, Lewes, DE) for seawater and oysters and B. J. Landau (Rutgers University, Multispecies Aquaculture Demonstration Facility, Cape May, NJ) for live algal cultures. We thank Ron Lau (Kona Coast Shellfish Co., Kailua-Kona, HI), William Burkhardt III (U.S. Food and Drug Administration, Dauphin Island, AL), Chris Langdon and Jonathan Sun (Hatfield Marine Science Center, Oregon State University, Newport, OR), and Sue Cudd (Whiskey Creek Shellfish Hatchery, Tillamook, OR) for collecting and shipping seawater samples. We acknowledge Mark Tamplin (University of Tasmania, Hobart, Australia) and Angelo DePaola (U.S. Food and Drug Administration, Dauphin Island, AL) for Vibrio cultures. We also thank David Kingsley (USDA, ARS, Dover, DE) and Joshua Gurtler (USDA, ARS, Wyndmoor, PA) for technical review of the manuscript.
This study was supported by the U.S. Department of Agriculture (USDA), Cooperative State Research, Education, and Extension Service (CSREES), National Research Initiative Grant 2008-35201-04535, by a USDA, Agricultural Research Service (ARS) Specific Cooperative Agreement 58-1935-0-043 with the University of Delaware, and by USDA, ARS intramural funds.
The use of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print 17 August 2012
REFERENCES
- 1. Beltrametti F, Kresse AU, Guzman CA. 1999. Transcriptional regulation of the esp genes of enterohemorrhagic Escherichia coli. J. Bacteriol. 181:3409–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boyd EF, et al. 2008. Molecular analysis of the emergence of pandemic Vibrio parahaemolyticus. BMC Microbiol. 8:110 doi:10.1186/1471-2180-8-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burnham JC, Hashimoto T, Conti SF. 1968. Electron microscopic observations on the penetration of Bdellovibrio bacteriovorus into gram-negative bacterial hosts. J. Bacteriol. 96:1366–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Centers for Disease Control and Prevention 1999. Outbreak of Vibrio parahaemolyticus infection associated with eating raw oysters and clams harvested from Long Island Sound - Connecticut, New Jersey, and New York, 1998. MMWR Morb. Mortal. Wkly. Rep. 48:48–51 [PubMed] [Google Scholar]
- 5. Chen H, Athar R, Zheng G, Williams HN. 2011. Prey bacteria shape community structure of their predators. ISME J. 5:1314–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen H, Williams HN. 2012. Sharing of prey: coinfection of a bacterium by a virus and a prokaryotic predator. mBio 3:e00051–12 doi:10.1128/mBio.00051-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chowdhury NR, et al. 2000. Molecular evidence of clonal Vibrio parahaemolyticus pandemic strains. Emerg. Infect. Dis. 6:631–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Daniels NA, et al. 2000. Emergence of a new Vibrio parahaemolyticus serotype in raw oysters: a prevention quandary. JAMA 284:1541–1545 [DOI] [PubMed] [Google Scholar]
- 9. Eksztejn E, Varon M. 1977. Elongation and cell division in Bdellovibrio bacteriovorus. Arch. Microbiol. 114:175–181 [DOI] [PubMed] [Google Scholar]
- 10. Fang FC, et al. 1992. The alternative σ factor KatF (RpoS) regulates Salmonella virulence. Proc. Natl. Acad. Sci. U. S. A. 89:11978–11982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Froelich BA, Williams TC, Noble RT, Oliver JD. 2012. Apparent loss of Vibrio vulnificus in North Carolina oysters coincides with drought-induced increase in salinity. Appl. Environ. Microbiol. 78:3885–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hengge-Aronis R. 2000. A role for the σ subunit of RNA polymerase in regulation of bacterial virulence. Adv. Exp. Med. Biol. 485:85–93 [DOI] [PubMed] [Google Scholar]
- 13. Hengge-Aronis R. 2002. Signal transduction and regulatory mechanisms involved in control of the sigmaS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hurley CC, Quirke A, Reen FJ, Boyd EF. 2006. Four genomic islands that mark post-1995 pandemic Vibrio parahaemolyticus isolates. BMC Genomics 7:104–122 doi:10.1186/1471-2164-7-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jones JL, et al. 2012. Biochemical, serological, and virulence characterization of clinical and oyster Vibrio parahaemolyticus isolates. J. Clin. Microbiol. 50:2343–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jones MK, Oliver JD. 2009. Vibrio vulnificus: disease and pathogenesis. Infect. Immun. 77:1723–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kadouri D, Venzon NC, O'Toole GA. 2007. Vulnerability of pathogenic biofilms to Micavibrio aeruginosavorus. Appl. Environ. Microbiol. 73:605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Konkel ME, Orfe L, Friel P, Call DR. 2012. Identification of potential type III secretion proteins via heterologous expression of Vibrio parahaemolyticus DNA. Appl. Environ. Microbiol. 78:3492–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kowarz L, Coynault C, Robbe-Saule V, Norel F. 1994. The Salmonella typhimurium katF (rpoS) gene: cloning, nucleotide sequence, and regulation of spvR and spvABCD virulence plasmid genes. J. Bacteriol. 176:6852–6860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lambert C, Morehouse KA, Chang C-Y, Sockett RE. 2006. Bdellovibrio: growth and development during predatory cycle. Curr. Opin. Microbiol. 9:639–644 [DOI] [PubMed] [Google Scholar]
- 21. Lambina VA, Afinogenova AV, Romay Z, Konovalova SM, Andreev LV. 1983. A new species of exoparasitic bacteria from the genus Micavibrio destroying gram-negative bacteria. Mikrobiologiia 52:777–780 [PubMed] [Google Scholar]
- 22. Lange R, Hengge-Aronis R. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5:49–59 [DOI] [PubMed] [Google Scholar]
- 23. Ma L, Chen J, Liu R, Zhang XH, Jiang YA. 2009. Mutation of rpoS gene decreased resistance to environmental stresses, synthesis of extracellular products and virulence of Vibrio anguillarum. FEMS Microbiol. Ecol. 70:130–136 [DOI] [PubMed] [Google Scholar]
- 24. Makino K, et al. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743–749 [DOI] [PubMed] [Google Scholar]
- 25. Motes ML, DePaola A. 1996. Offshore suspension relaying to reduce levels of Vibrio vulnificus in oysters (Crassostrea virginica). Appl. Environ. Microbiol. 62:3875–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Motes ML, et al. 1998. Influence of water temperature and salinity on Vibrio vulnificus in Northern Gulf and Atlantic Coast oysters (Crassostrea virginica). Appl. Environ. Microbiol. 64:1459–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nair GB, et al. 2007. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20:39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nielsen AT, et al. 2006. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2:e109 doi:10.1371/journal.ppat.0020109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nishibuchi M, Kaper JB. 1995. Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect. Immun. 63:2093–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Okuda J, et al. 1997. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35:3150–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Park DK, et al. 2006. Cyclo(Phe-Pro) modulates the expression of ompU in Vibrio spp. J. Bacteriol. 188:2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pineiro SA, et al. 2007. Global survey of diversity among environmental saltwater Bacteriovoracaceae. Environ. Microbiol. 9:2441–2450 [DOI] [PubMed] [Google Scholar]
- 33. Richards GP, Hammer CH, Garfield MK, Parveen S. 2004. Characterization of a lysyl aminopeptidase activity associated with phosphoglucose isomerase of Vibrio vulnificus. Biochim. Biophys. Acta 1700:219–229 [DOI] [PubMed] [Google Scholar]
- 34. Rittenberg SC, Shilo M. 1970. Early host damage in the infection cycle of Bdellovibrio bacteriovorus. J. Bacteriol. 102:149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rosche TM, Smith DJ, Parker EE, Oliver JD. 2005. RpoS involvement and requirement for exogenous nutrient for osmotically induced cross protection in Vibrio vulnificus. FEMS Microbiol. Ecol. 53:455–462 [DOI] [PubMed] [Google Scholar]
- 36. Starr MP, Baigent NL. 1966. Parasitic interaction of Bdellovibrio bacteriovorus with other bacteria. J. Bacteriol. 91:2006–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tison DL. 1999. Vibrios, p 497–506 In Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH. (ed), Manual of clinical microbiology, 7th ed ASM Press, Washington, DC [Google Scholar]
- 38. Tudor JT, McCann MP, Acrich IA. 1990. A new model for the penetration of prey cells by bdellovibrios. J. Bacteriol. 172:2421–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Typas A, Becker G, Hengge R. 2007. The molecular basis of selective promoter activation by the sigmaS subunit of RNA polymerase. Mol. Microbiol. 63:1296–1306 [DOI] [PubMed] [Google Scholar]
- 40. Vasudevan P, Venkitanarayanan K. 2006. Role of the rpoS gene in the survival of Vibrio parahaemolyticus in artificial seawater and fish homogenate. J. Food Prot. 69:1438–1442 [DOI] [PubMed] [Google Scholar]
- 41. Reference deleted.
- 42. Whitaker WB, Parent MA, Boyd A, Richards GP, Boyd EF. 2012. Vibrio parahaemolyticus ToxRS regulator is required for stress tolerance and colonization in a novel orogastric streptomycin-induced adult murine model. Infect. Immun. 80:1834–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Whitaker WB, et al. 2010. Growth of Vibrio parahaemolyticus O3:K6 at different salt concentrations modulates responses to pH and temperature stresses. Appl. Environ. Microbiol. 76:4720–4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Williams HN, Baer ML. 2005. Genus II bacteriovorax, p 1053–1057 In Garrity GM, Brenner DJ, Krieg NR, Staley JT. (ed), Bergey's manual of systematic bacteriology: the Proteobacteria, vol 2, part C Springer, New York, NY [Google Scholar]
- 45. Williams HN, Baer ML, Tudor JJ. 2005. Bdellovibrio Stolp and Starr, p 1040–1053 In Garrity GM, Brenner DJ, Krieg NR, Staley JT. (ed), Bergey's manual of systematic bacteriology: the Proteobacteria, vol 2, part C Springer, New York, NY [Google Scholar]
- 46. Williams HN, Falker WA, Jr, Shay DE. 1980. Incidence of marine bdellovibrios lytic against Vibrio parahaemolyticus in Chesapeake Bay. Appl. Environ. Microbiol. 40:970–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Williams HN, Falker WA, Jr, Shay DE. 1982. Seasonal distribution of bdellovibrios at the mouth of the Patuxent River in the Chesapeake Bay. Can. J. Microbiol. 28:111–116 [DOI] [PubMed] [Google Scholar]
- 48. Williams HN, Turng B-F, Kelley JI. 2009. Survival response of Bacteriovorax in surface biofilm versus suspension when stressed by extremes in environmental conditions. Microb. Ecol. 58:474–484 [DOI] [PubMed] [Google Scholar]