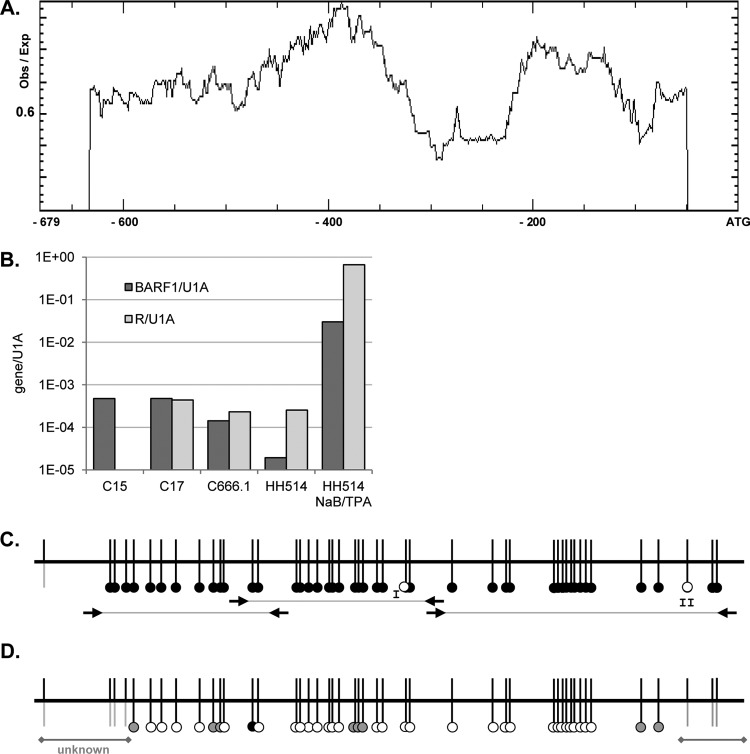
Fig 1.
Methylation of the BARF1 promoter region. (A) Methprimer analysis of CpG islands in the BARF1 promoter showed two large CpG islands spanning the whole promoter up to −607 nucleotides relative to the BARF1 ATG start site. Obs, observed; Exp, expected. (B) Quantitative PCR of BARF1 and R mRNA normalized to U1A. BARF1 mRNA was detected both in C15 and C17 NPC mouse tumor material and in C666.1 NPC cells. R levels in C15 mouse tumor material were below the detection limit. BARF1 mRNA was present at low levels in HH514 cells but could be expressed upon treatment with NaB and TPA. (C) Bisulfite sequencing PCR (BSP) using primers as indicated (arrows) was performed to analyze the CpG islands on the BARF1 promoter from ATG-632 to the ATG start site. Almost all CpGs in the investigated samples were methylated (black dots), both in HH514 Burkitt's lymphoma cells and in C666.1 cells consistently harboring EBV and C15 and C17 mouse xenograft NPC tumor material. CpG site I was unmethylated (white dots) in C666.1 cells and C17 tumor material. CpG site II was unmethylated in HH514 cells. (D) BSP analysis of 24-h NaB-TPA-treated HH514 cells shows that the BARF1 promoter was partially (gray dots) or completely (white dots) demethylated.