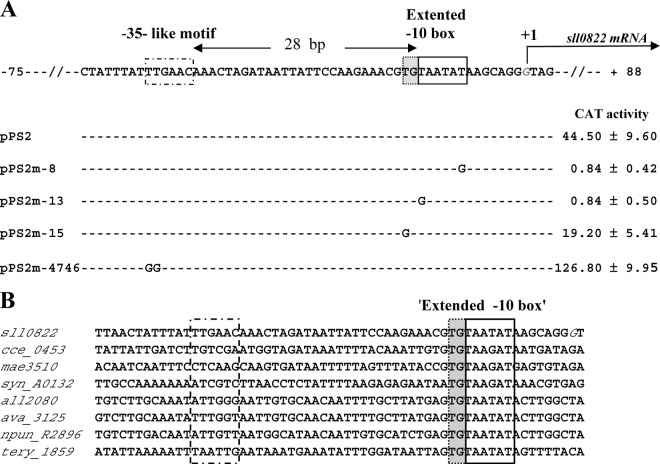
Fig 3.
Mutational analysis in Synechocystis of the abrB2 promoter transcriptionally fused to the cat reporter gene of our replicative promoter probe vector. (A) Sequence alignment of the nontranscribed DNA strand of the WT (pPS2 reporter plasmid) and mutant (pPS2m reporter plasmids) abrB2 promoters. Sequence resembling the −35 and extended −10 promoter elements are boxed, and the origin of transcription (+1; G nucleotide in italics) that we mapped in this study (see Fig. S1 in the supplemental material) is indicated by the bent arrow. Nucleotide substitutions in the mutant promoters (m-8, m-13, m-15, and m-4746) are shown in uppercase letters, while the conserved nucleotides are indicated by dashed lines. For each reporter strain, the CAT activity is the mean value of three measurements performed on two independent cellular extracts; 1 CAT unit = 1 nmol of chloramphenicol acetylated/min/mg of protein. (B) Sequence alignments of the abrB2 promoter regions from various cyanobacteria resembling Synechocystis in that their abrB2 gene is divergently transcribed with an opposite gene. Besides Synechocystis PCC6803 (sll0822), these cyanobacteria are as follows: Cyanothece ATCC 51142 (cce_0453), Microcystis aeruginosa NIES-843 (mae_35130), Nostoc PCC 7120 (all2080), Anabaena variabilis ATCC 29413 (ava_3125), Nostoc punctiforme PCC 73102 (npun_R2896), and Trichodesmium erythraeum ISM101 (tery_1859). The positions of the conserved extended −10 promoter element and of the nonconserved sequence resembling a −35 element in Synechocystis are indicated by boxes.