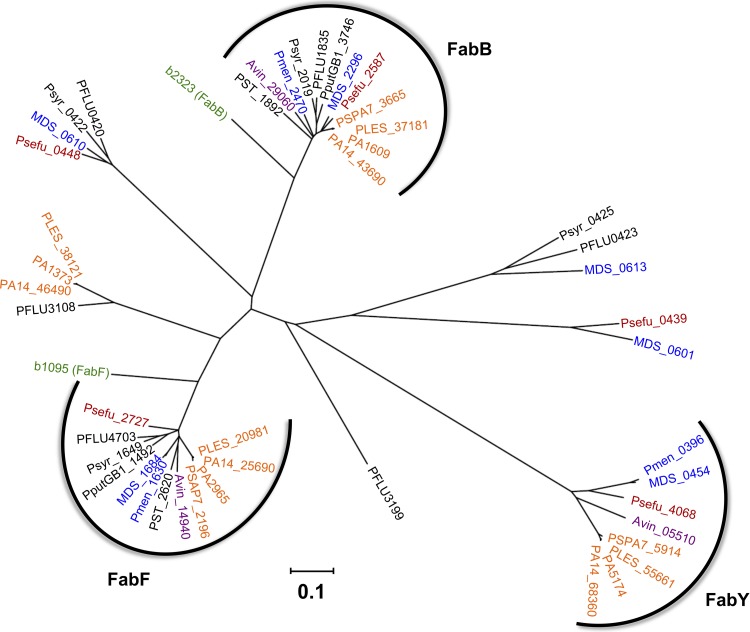
Fig 9.
Phylogenetic tree and distribution of KASI/II domain-containing proteins among pseudomonads. All open reading frames within each genome having significant similarity to P. aeruginosa PAO1 KASI/II proteins (E-value < 1e−10) and that are not a subdomain of a polyketide synthase multidomain protein were included in the analysis. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The tree is drawn to scale, with branch lengths in the units of number of amino acid substitutions per site. Analyses were conducted by using MEGA5 (82). The three main KASI/II groups (FabB, FabF, and FabY) are indicated, and all members within each cluster share at least partial genomic synteny. Locus tag abbreviations: PA, P. aeruginosa PAO1 (orange); PSPA7, P. aeruginosa PA7 (orange); PA14, P. aeruginosa UCBPP-PA14 (orange); PLES, P. aeruginosa LESB58 (orange); Pmen, P. mendocina ymp (blue); MDS, P. mendocina NK-01 (blue); Psefu, P. fulva 12-X (red), PST, P. stutzeri A1501 (black), PputGB1, P. putida GB-1 (black); Psyr, P. syringae pv. syringae B728a (black); PFLU, P. fluorescens SBW25 (black); Avin, Azotobacter vinelandii DJ (purple); b, E. coli (green).