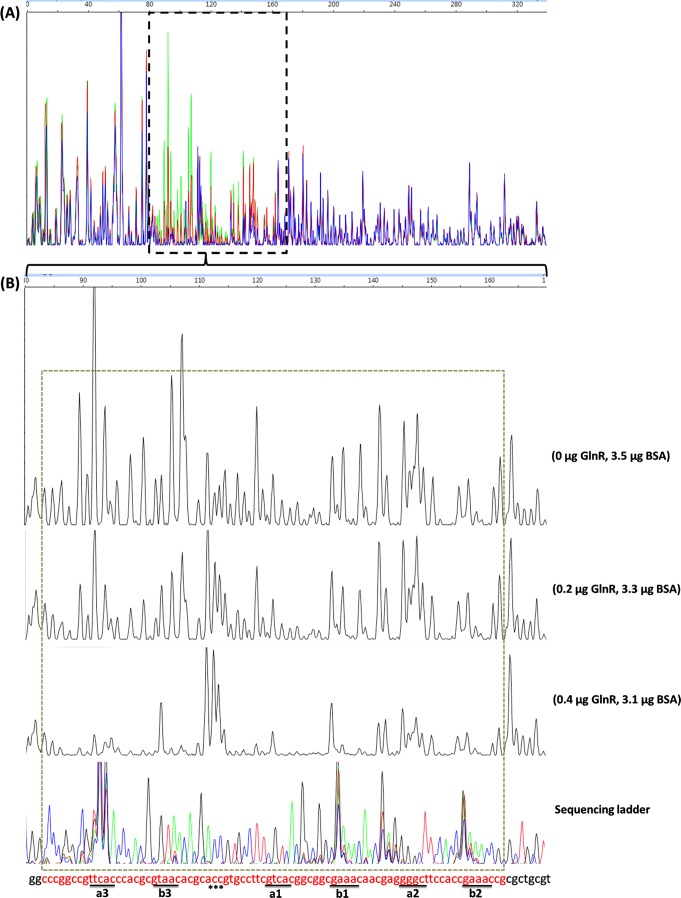
Fig 1.
Identification of the GlnR-protected cis-elements in the S. coelicolor amtB promoter region using a DNase I footprinting assay. The probes used were labeled with FAM dye and are described in Materials and Methods. The whole region of the coding strand DNA is shown in panel A, and the region protected by GlnR from DNase I cleavage was enlarged and is shown separately in panel B. In panel A, the colored lines represent the different concentrations of GlnR used: green, 0 μg; red, 0.2 μg; and blue, 0.4 μg. In panel B, the GlnR binding boxes are underlined, and the DNase I-hypersensitive sites (ACC) are denoted by asterisks. The protected patterns remained the same even when the amount of GlnR used for the binding assay increased up to 2.4 μg (data not shown). The fmol DNA cycle sequencing system was used for DNA sequencing reactions. The sequencing products, together with the digestion products, were analyzed with an ABI 3130xl DNA analyzer and peak scanner software v1.0 (Applied Biosystems). The four sequencing results (G, A, T, and C) are indicated by four different colors separately and are then merged together. The overlapping peaks were probably caused by higher secondary structures within the promoter region. The electropherograms were aligned together with the use of standards. BSA, bovine serum albumin.