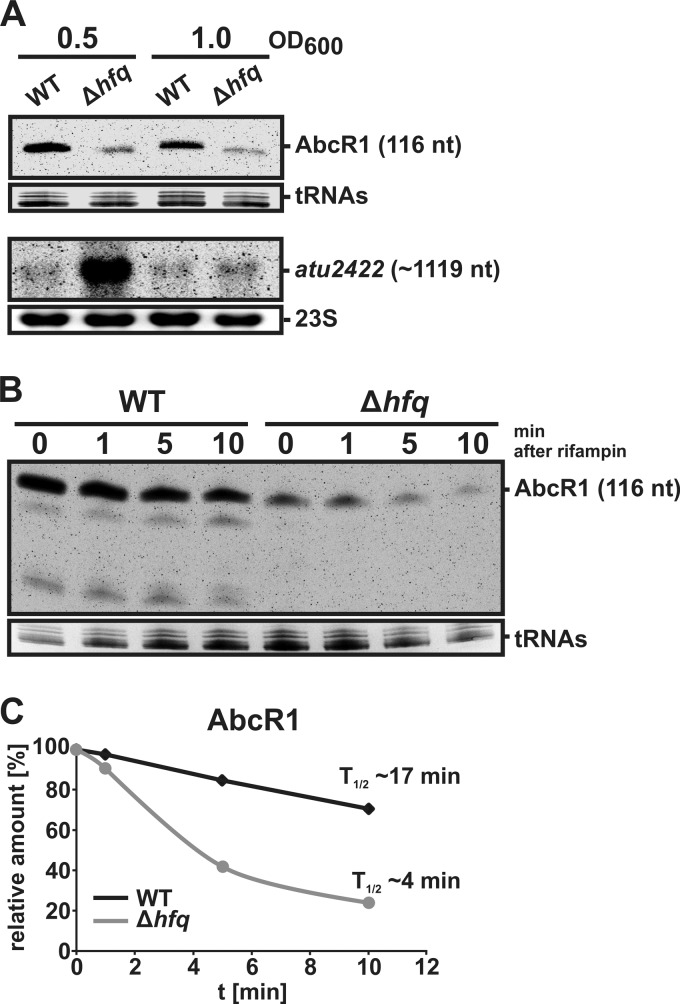
Fig 2.
Effect of Hfq on AbcR1-mediated gene regulation in vivo. (A) Northern blot analysis of AbcR1 and atu2422 in A. tumefaciens wild type (WT) and the Δhfq strain. Hybridizations were performed with 8 μg of total RNA from cells grown in YEB complex medium to the optical densities indicated above the gels. Primers used for RNA probe generation are listed in Table S2 in the supplemental material. Ethidium bromide-stained 23S RNAs or tRNAs were used as loading controls. nt, nucleotides. (B) Stability of AbcR1. WT and Δhfq mutant cells were grown to an OD600 of 1.0 in YEB medium. Samples were taken before (0) and 1, 5, and 10 min after the addition of rifampin (250 μg/ml). Amounts of 8 μg of total RNA were separated on a 10% polyacrylamide gel containing 7 M urea and were detected by Northern analysis using a DNA probe (generated by primers DNAprobe_C2A_fw and DNAprobe_C2A_rv). The size of AbcR1 is given on the right. Ethidium bromide-stained tRNAs were used as loading controls. (C) Relative amounts of AbcR1 in A. tumefaciens WT and the hfq mutant after the addition of rifampin. Northern blot signals (B) were quantified using AlphaEaseFC software.