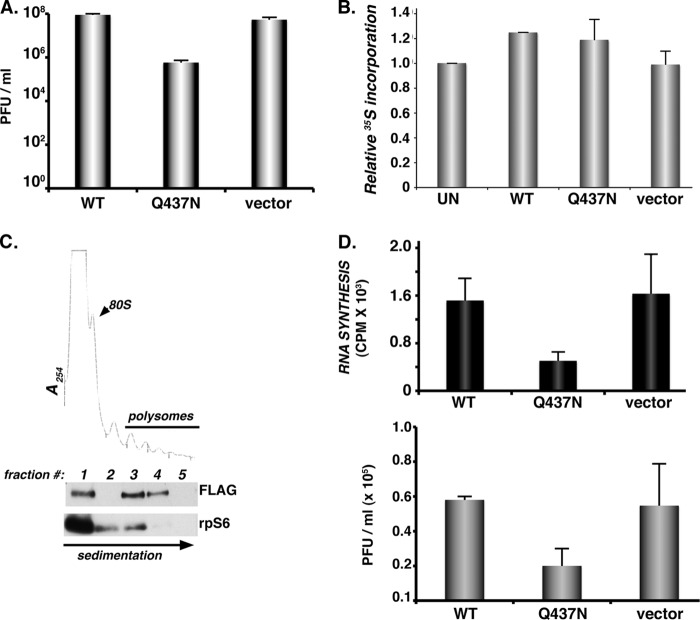
Fig 6.
Site-specific PABP cleavage stimulates EMCV RNA synthesis and infectious virus production. (A) HeLa cells transfected with plasmids expressing WT PABP, the Q437N cleavage-resistant variant (as in the Fig. 4D legend), or the plasmid vector-only control were infected with EMCV (MOI = 10−3). After 48 h, lysates were prepared by freeze-thawing, and the amount of infectious virus produced was quantified by plaque assay in Vero cells. (B) HeLa cells were transiently transfected as for panel A, transfected with an EGFP-expressing plasmid, or left untransfected. After 48 h, cultures were metabolically labeled for 1 h as for panel A. Total protein was isolated and the amount of 35S incorporated determined by trichloroacetic acid (TCA) precipitation. Acid-insoluble material was collected onto fiberglass filters and washed, and the radioactivity was quantified by counting in liquid scintillant. The relative amount of 35S incorporation in untransfected HeLa cells was normalized to 100%. Error bars indicate the standard errors of the means. (C) Cell-free lysates prepared from HeLa cells ectopically expressing cleavage-resistant Flag-tagged PABP variant (Q437N) were loaded onto sucrose gradients, which were subsequently fractionated while absorbance at 254 nm was monitored. TCA-precipitated fractions were analyzed by immunoblotting using antibodies specific for Flag or ribosomal protein s6 (rpS6). Fraction 1, free ribosome subunits; fraction 2, disomes; fractions 3 to 5, polyribosomes. (D) HeLa cells transiently transfected with empty plasmid vector or a plasmid expressing Flag-tagged PABP (WT or Q437N) were infected with EMCV (MOI = 10) for 5 h. (Top) Infections were performed in the presence of [3H]uridine and actinomycin D. Total RNA was isolated, samples were precipitated with TCA, and the amount of acid-insoluble radioactivity quantified by counting in liquid scintillant. Errors bars indicate standard errors of the means. (Bottom) The amount of infectious virus produced after a single replication cycle was quantified by plaque assay. Error bars indicate standard errors of the means.