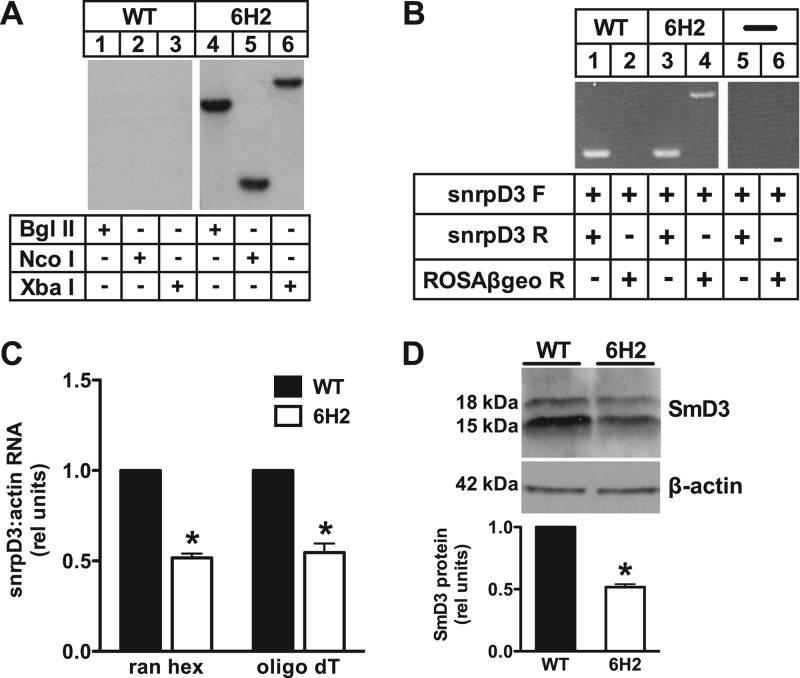
Fig 2.
6H2 cells are haploinsufficient for SmD3. (A) Autoradiogram showing Southern blot analysis of WT (lanes 1 to 3) and 6H2 (lanes 4 to 6) genomic DNA digested with the restriction enzyme BglII, NcoI, or XbaI. The blot was probed with a 32P-labeled fragment corresponding to the ROSAβgeo sequence. (B) PCR was performed on cDNA from WT (lanes 1 and 2) and 6H2 (lanes 3 and 4) cells with reaction mixtures containing no cDNA as controls (lanes 5 and 6). Forward (F) and reverse (R) primers for snrpD3 were designed to detect endogenous snrpD3 (lanes 1, 3, and 5). The forward snrpD3 primer and reverse primer for the proviral sequence were used to detect fusion transcript (lanes 2, 4, and 6). (C) RNA was isolated from WT and 6H2 cells and reverse transcribed using either random hexamers (ran hex) to prime total RNA or oligo(dT) to prime mRNA. snrpD3 expression was determined by qRT-PCR and normalized to β-actin expression. (D) Protein expression in WT and 6H2 cells was determined by Western blotting and quantified by densitometry (bands at 15 kDa and 18 kDa likely reflect known posttranslational modification of SmD3). A representative blot is shown for SmD3 and β-actin control. On the bar graphs, the data are expressed as means and SE for three independent experiments. *, P < 0.05 for 6H2 versus WT.