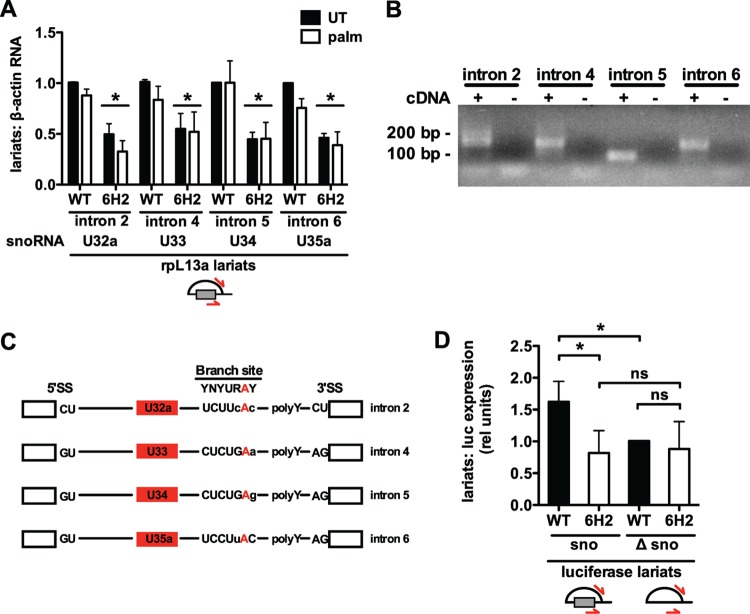
Fig 8.
snoRNA-containing intron lariats are decreased in 6H2 cells. (A) WT and 6H2 CHO cells were untreated or treated with palm for 9 h. Nuclear RNA was isolated and reverse transcribed using random hexamers. rpL13a intron lariat abundance was determined by qRT-PCR using primers reading across branch points. Quantification of the lariat PCR product was normalized to β-actin mRNA. (B and C) PCR products generated by intron lariat primers in CHO WT cells were visualized on a 2% agarose gel (B) or cloned and sequenced (C). (B) Lanes with cDNA (+) contained reverse transcription product as the template. Lanes without cDNA (−) were negative-control PCRs with no reverse transcription product provided as the template. (C) Sequencing revealed the branch site adenosine (red) for each intron. Branch site nucleotides showing conservation (uppercase) are indicated. (D) WT and 6H2 cells were transfected with a split luciferase construct containing the intact murine U32a intron (sno) or the U32a intron lacking the 83-nucleotide U32a snoRNA (Δsno). Intron lariat abundance was determined by qRT-PCR using primers specific for the murine lariat sequences and normalized to luciferase pre-mRNA expression. All data are expressed as means and SE for three independent experiments. *, P < 0.05; ns, not significant.