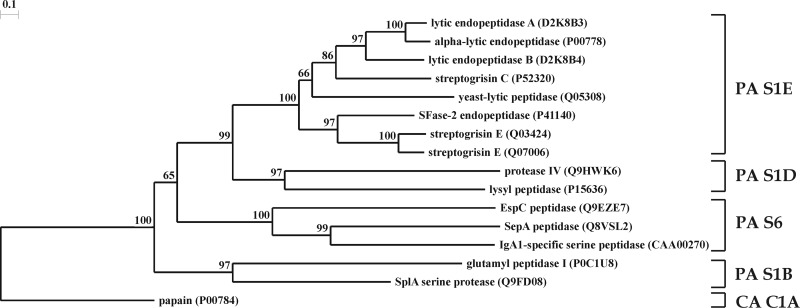
Fig 2.
Phylogeny of endopeptidases AlpA and AlpB. This phylogenetic tree of bacterial serine proteases was constructed on the basis of homology between the amino acid sequences of mature enzymes. Accession numbers of sequences in the UniProt database are given. Enzymes from the following organisms are presented: P00778, L. enzymogenes; D2K8B3 and D2K8B4, Lysobacter sp. strain XL1; P52320 and Q07006, Streptomyces griseus; Q03424 and P41140, Streptomyces fradiae; Q05308, Rarobacter faecitabidus; P0C1U8 and Q9FD08, S. aureus; P15636, Achromobacter lyticus; Q9HWK6, Pseudomonas aeruginosa PAO1; Q8VSL2, Shigella flexneri; CAA00270 (GenBank accession number), Neisseria gonorrhoeae; Q9EZE7, E. coli; P00784, Carica papaya. Papain was selected as an outgroup. Protease clans (PA and CA) and families (S1E, S1B, S1D, and S6) are given at the right in accordance with the MEROPS peptidase database (34; http://merops.sanger.ac.uk/).