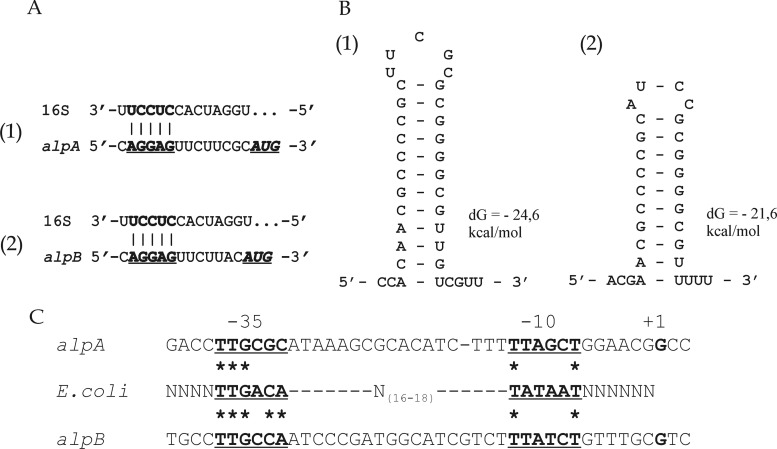
Fig 5.
Analysis of translation and transcription signals for alpA (parts 1) and alpB (parts 2) of Lysobacter sp. strain XL1. (A) Fragments of the 5′ UTR sequences of the alpA (bp 2391 to 2407) and alpB (bp 4190 to 4205) mRNAs and the sequence at the 3′ end of the 16S rRNA of Lysobacter sp. strain Shinshu-th3 (bp 1530 to 1543; GenBank accession no. AB121774) are shown. The putative ribosome-binding sites in the mRNAs of alpA and alpB are in bold and underlined. The start codons are in bold italics and underlined. (B) Fragments of the 3′ UTR sequences of the alpA (bp 3628 to 3656) and alpB (bp 5430 to 5453) mRNAs capable of forming stem-loop structures are shown. The sequences were analyzed using the UNAFold package (27; http://mfold.rna.albany.edu/). Hairpin energy (in kcal/mol) is given on the right. Positions of the ribosome-binding sites and stem-loop structures are given in accordance with GenBank accession no. GU188567. (C) Analysis of alpA and alpB −35 and −10 regions compared with the corresponding consensus sequences of the E. coli σ70 promoter (bold and underlined). The transcription start point (+1) is in bold. Asterisks indicate identical bases.