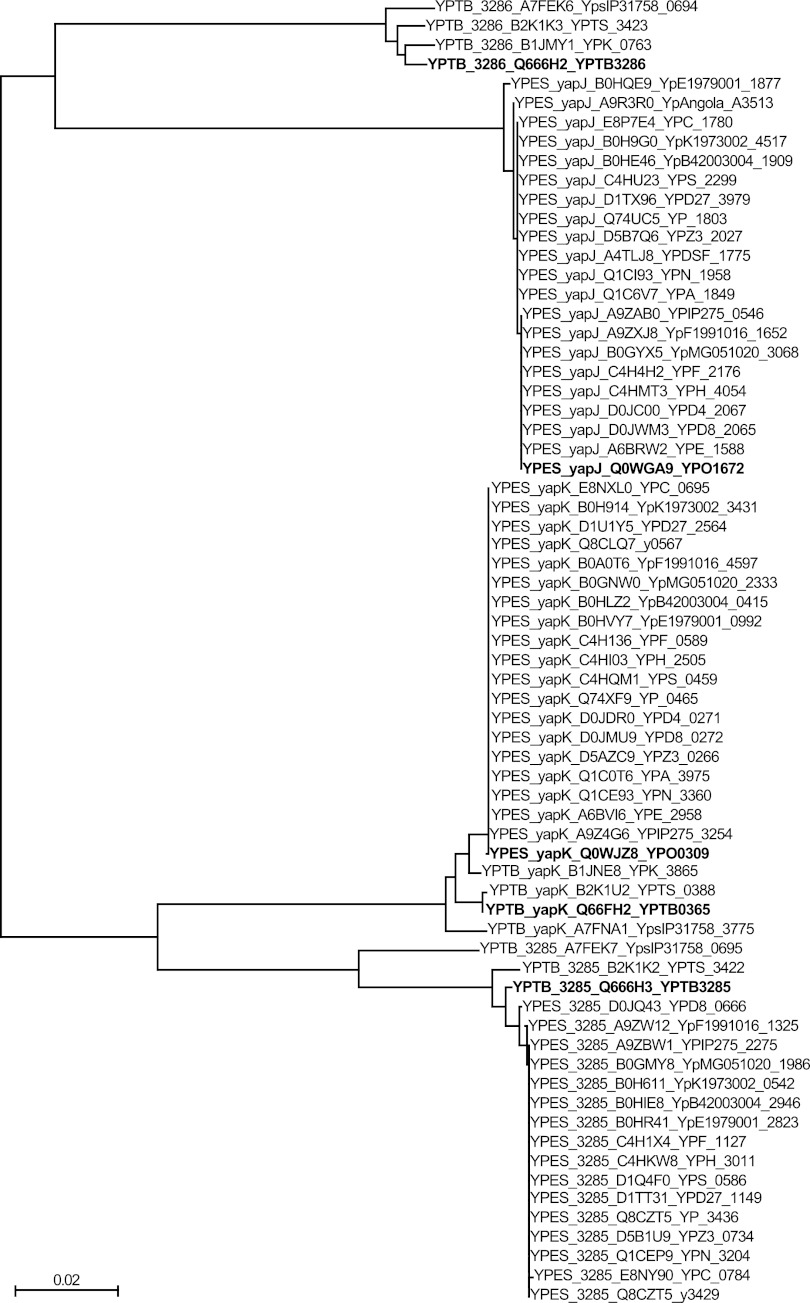
Fig 2.
Neighbor-joining phylogenetic tree of YapJ- and YapK-like protein sequences. Sixty-eight genes predicted to produce protein products similar to YapJ, YapK, YPTB0365, YPTB3285, or YPTB3286 were identified in sequenced genomes of Y. pestis and Y. pseudotuberculosis available through the NCBI. Translated sequences were aligned by using MUSCLE and used to generate a bootstrapped phylogenetic tree (ClustalX v. 2.0). Major nodes defining the phylogenetic relationships between the YapJ, YapK, YPTB3285, and YPTB3286 protein families were found 100% of the time in the bootstrapped trees. Proteins are listed first by their species of origin, then by the reference protein to which they are closest (YapJ, YapK, YPTB3285, or YPTB3286), then by their UniProt identification, and finally by their NCBI accession number. YPTB0365 falls within the same group as YapK due to their extremely high level of identity. The proteins investigated in this study (YapJ and YapK from Y. pestis CO92 and YPTB0365, YPTP3285, and YPTB3286 from Y. pseudotuberculosis IP32953) are identified in boldface type for ease of reference. YPES, Y. pestis sequences; YPTB, Y. pseudotuberculosis sequences. The scale bar indicates the number of amino acid substitutions per site for each branch.