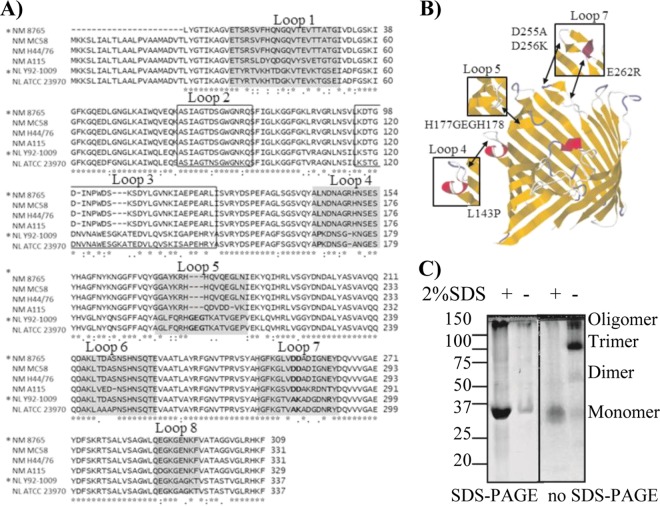
Fig 1.
PorB sequence comparison and analysis of PorB mutants. (A) Alignment of PorB sequences from N. meningitidis serogroup B strains 8765 (*) (homologous to H44/76), MC58, and H44/76 and serogroup A strain A115 and N. lactamica strains Y92-1009 (*) and ATCC 23970, performed with the ClustalW2 alignment tool. Loop 1, 4, 5, 6, 7, and 8 are shaded. Loops 2 and 3 are boxed. Residues in loops 4, 5, and 7 chosen for mutagenesis are in bold. (B) Theoretical model of PorB monomer (N. meningitidis strain 8765) based on the N. meningitidis PorB template (PBID 3A2R) (49) created with EasyPre3D server (21a) using RasMol (43a) and Jalview (9a, 55a). The position of mutated residues in loops 4, 5, and 7 is indicated by the arrows, and the effect on the corresponding loop regions is indicated in the insets. (C) Coomassie staining of wtPorB, representative of purified recombinant PorB molecules. (Left) Conventional SDS-PAGE; (right) modified PAGE (SDS free). Prior to electrophoresis, PorB aliquots were heated in sample loading buffer in the presence or absence of SDS, as indicated. Bands with molecular weights corresponding to monomers, dimers, trimers, and oligomers were observed.