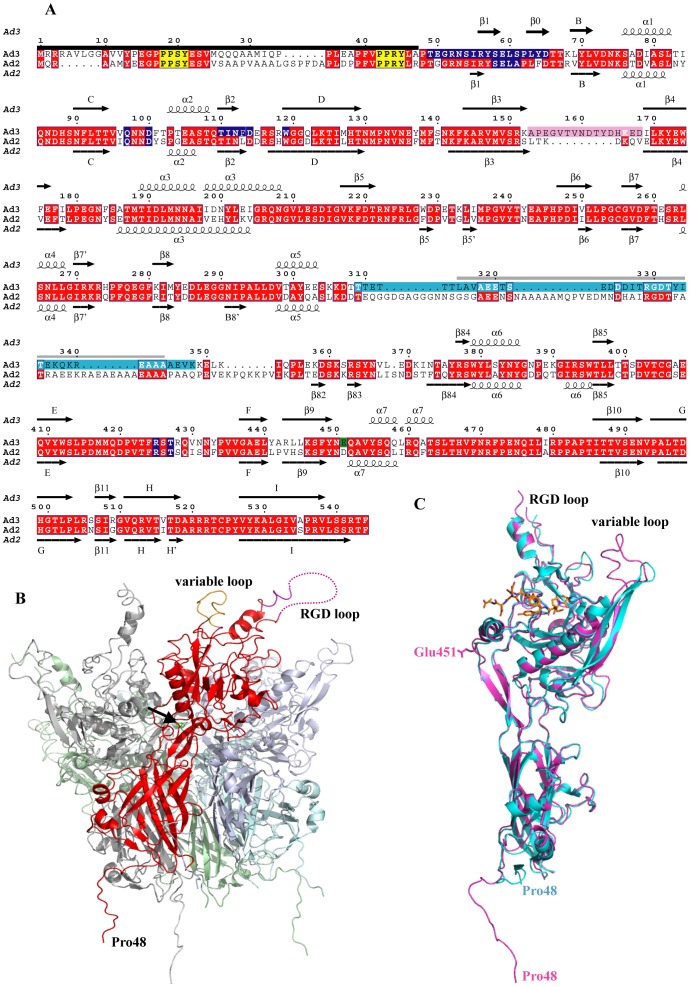
Figure 1. Ad3 dodecahedron structure.
(A) The secondary structure of the Ad3 Pb in the orthorhombic crystal form is shown together with the one of Ad2 (pdb entry 1×9p [13]). The corrected sequence (GenBank acc. no ABB17799.1 [26]) of the Ad3 Pb with 58SELS instead of 58SDVS has been used. Disordered or proteolysed residues at the N-terminus which are invisible in the structure are indicated with a black bar. PPxY motifs are marked in yellow. Glu451 involved in a putative Ca2+ binding site is highlighted with a green background, the variable loop with a pink background, the RGD loop with a light blue background. The peptide of the RGD binding loop that is probably missing due to proteolysis, is marked with a gray bar. A dark blue background marks contact residues involved in the Dd formation. Secondary structure elements are labeled as in Zubieta et al. [13] whenever possible. (B) Cartoon representation of the overall structure of an Ad3 Pb in the cubic crystal form. One of the Pb subunits is highlighted in red. The dotted line symbolizes the part of the RGD loop, which is invisible in electron density. The black arrow points to the putative calcium ion. The ‘dangling’ N-terminal ends at the bottom are in fact involved in strand-swapping with other Pbs. (C) Alignment of the Ad3 Pb monomer structure (magenta) from the orthorhombic crystal form in presence of fiber peptide (orange) with the one of Ad2 (cyan).