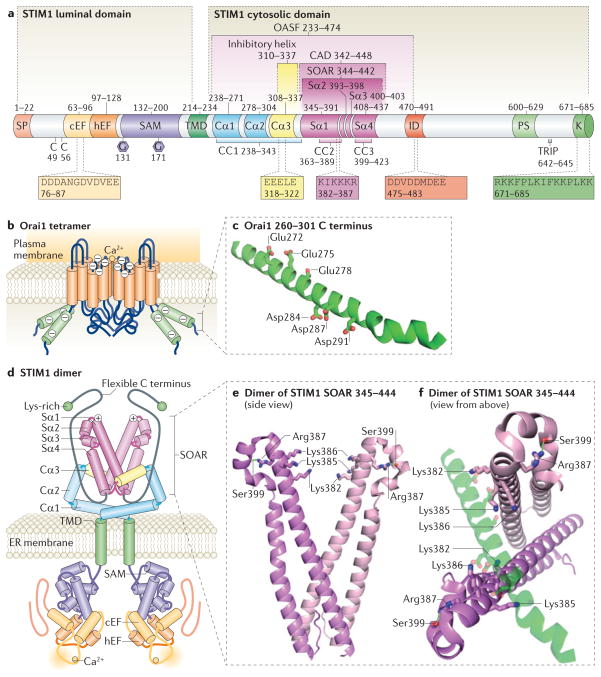
Figure 1. Structure and activation of STIM1.
a| The molecular domains of stromal interaction molecule 1 (STIM1). Endoplasmic reticulum (ER) STIM1 contains a luminal and a cytosolic domain. The amino-terminal signal peptide (SP) is cleaved during translation. The ER luminal N-terminal domain includes a conserved Cys pair, a Ca2+-binding canonical EF-hand domain, (cEF), a non-Ca2+-binding hidden EF-hand (hEF) domain, a sterile α-motif (SAM) with two Asn-linked glycosylation sites (shown as hexagons) and a single transmembrane domain (TMD). The cytosolic carboxy-terminal domain is considered to include three coiled-coil regions74 (called CC1, CC2 and CC3). CC1 is divided into three α-helices (termed Cα1, Cα2 and Cα3) on the basis of sequence analysis predictions by using JPred3 (REF. 188). The structure of Cα3 is also determined on the basis of homology with the recently solved Caenorhabditis elegans STIM structure40. SOAR (STIM–Orai activating region) is the minimal sequence required for the activation of Orai1 (REF. 45). SOAR contains four α-helices, termed Sα1, Sα2, Sα3 and Sα4 (REF. 40). The segments CAD (Ca2+ release-activated Ca2+ (CRAC) activation domain)46 and OASF (Orai1-activating small fragment)47 are larger than SOAR, contain the CC1 region and also activate Orai1. SOAR includes the polybasic region, with the sequence KIKKKR (amino acids 382–387), which is crucial for the interaction with Orai1 (REFS 40,68,71). Cα3 contains an inhibitory helix that inhibits SOAR function40,68,71. The acidic EEELE (residues 318–322) region is required for the action of the inhibitory helix40,68. Downstream of SOAR resides an acidic inhibitory domain (ID) that mediates fast Ca2+-dependent inactivation of Orai1 (REFS 89,92,93). The C-terminal tail contains a Pro/Ser-rich domain (PS), a microtubule interacting domain (TRIP) and a Lys-rich domain responsible for phospholipid interaction at the plasma membrane. b | The tetrameric structure of the Orai1 channel is shown, highlighting the TMDs (shown in orange), extracellular and intracellular sequences (blue) and C-terminal predicted α-helices (shown in green). Negatively charged residues are indicated in the C-terminal helices and in the Ca2+ selectivity filter at the predicted mouth of the pore. c | The predicted α-helical structure of the Orai1 C terminus and potential sites for SOAR binding are illustrated on the basis of secondary structure prediction by using JPred3 (REF. 188). Side chains from acidic residues Glu272, Glu275, Glu278, Asp284, Asp287 and Asp291 are shown as amphipathic helices that may electrostatically interact with basic residues in the SOAR dimer. d | Proposed structure of a resting STIM1 dimer. The predicted α-helices in CC1 are indicated in the figure and are shown in a folded configuration. The inhibitory Cα3 helix (highlighted in yellow) is bound to SOAR (shown in red), which comprises four α-helices as indicated. The SOAR polybasic region is shown (+). The STIM1 dimer is held together predominantly by interactions between the CC1 and SOAR regions37,40. The C-terminal flexible region (shown in grey) together with the C-terminal Lys-rich domain (shown in green) may provide some steric shielding of SOAR77. α-helices are shown as cylinders; flexible regions as lines. e | The side view of the SOAR structure reveals that Lys382, Lys385 and Lys386 are located in the polybasic Orai-interacting region, and that these residues are oriented towards the centre of the cleft. Lys384 (not shown) is oriented in another direction. The Arg387 residue may be involved in hydrogen bonding with Ser399 (indicated by a green line). Coordinates were obtained from Protein data bank entry 3TEQ40. f | SOAR structure shown in (e) rotated 90° to illustrate the potential sites of association with the Orai1 C terminus depicted in (c). The hypothetical electrostatic binding of the Orai1 C terminus within the cleft between the SOAR monomers is shown.