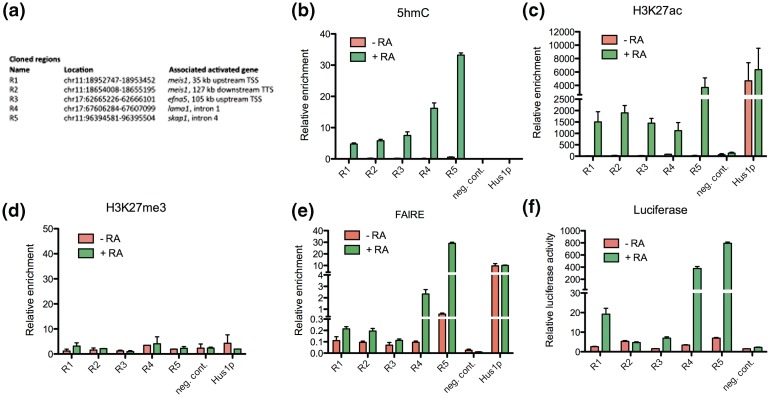
Figure 4.
Regions defined by coordinated variations in 5hmC, H3K4me2 and H3K27ac in P19 cells are RA-dependent enhancers. (a) Table indicating the genomic coordinates of selected 5hmC-up regions cloned in a luciferase reporter vector and their position relative to RA-regulated genes. (b) Validation of 5hmC variations in selected regions by hMeDIP-qPCR shown as relative enrichment (percentage of input). (c) ChIP-qPCR analysis of H3K27ac enrichment of selected regions. The open chromatin of the Hus1 promoter served as a positive control, whereas amplification of a region within Nf1a on chromosome 4 provided a negative control. Results are shown as mean ± SEM of biological duplicates. (d) ChIP-qPCR analysis of H3K27me3 enrichment of selected regions. Negative and positive controls are as in (c). Results are shown as mean ± SEM of biological duplicates. (e) FAIRE assay of nucleosome depletion/stability in selected regions. Results are shown as mean ± SEM of biological triplicates. (f) Luciferase reporter assay of the cloned regions −/+ RA. Negative control indicates relative luciferase activities obtained after transfection of the empty control vector.