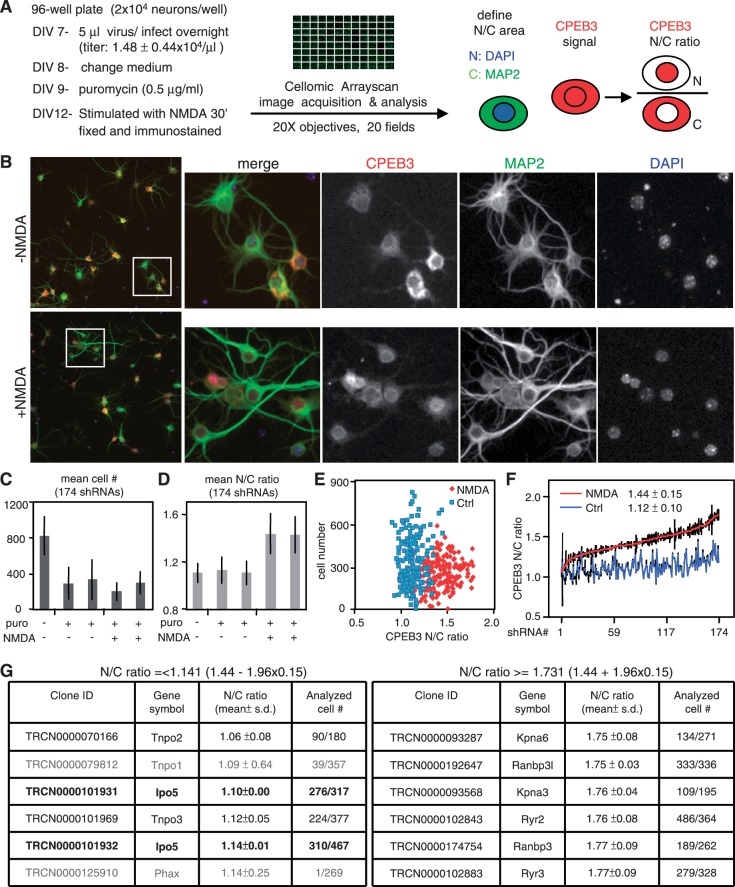
Figure 3.
Identification of the trans-acting molecules affecting nucleocytoplasmic distribution of CPEB3. (A) The scheme of virus-based shRNA knockdown screening to identify candidate karyopherins (see Supplementary Table S1 for all shRNA information). The neurons infected with lentiviruses expressing 174 shRNAs were stimulated with or without 20 µM NMDA and detected with MAP2 and CPEB3 antibodies and DAPI nuclear staining. N: nucleus; C: cytoplasm. Representative images from the screening are shown in (B). (C) The average cell number and (D) CPEB3 N/C ratio in each dataset (calculated from 174 shRNA’s data) are displayed as bar graphs. (E) The cell numbers in each shRNA-infected well from duplicate datasets were averaged and plotted against the mean CPEB3 N/C ratio. (F) The mean CPEB3 N/C ratios obtained from 174 shRNA-targeted and NMDA-treated neurons are arranged in an increasing order (red line) and the ratios of their untreated counterparts are plotted in blue. The average CPEB3 N/C ratios from Ctrl- and NMDA-treated datasets are 1.12 ± 0.10 and 1.44 ± 0.15 (mean ± SD), respectively. (G) The shRNAs, which affect the CPEB3 N/C ratio beyond 95% confidence interval (mean ± 1.96 SD) in NMDA-stimulated neurons are listed. All error bars denote SD.