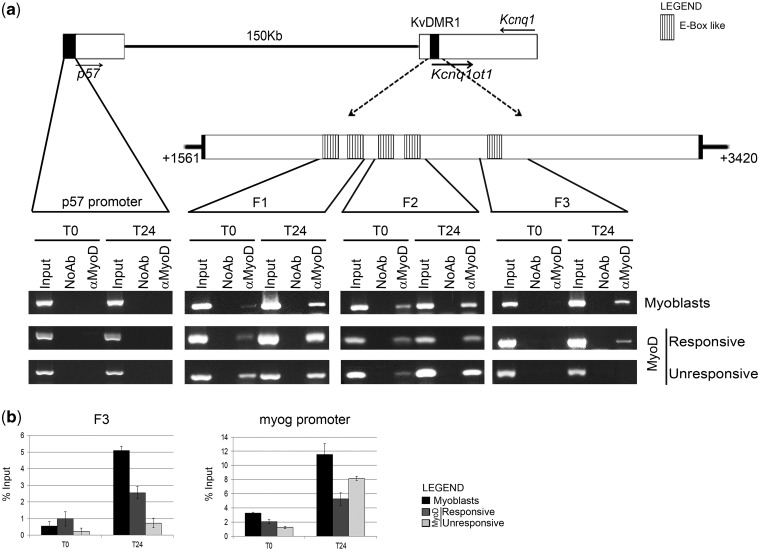
Figure 2.
MyoD interacts in vivo with KvDMR1. (a) Top: schematic diagram showing the reciprocal location of p57, Kcnq1 and Kcnq1ot1 genes and KvDMR1 region. Filled arrows indicate the direction of transcription. Dashed arrows point to an enlargement of the repressive KvDMR1 element previously described (31) extending from nt +1561 to +3420 of AF119385 sequence. The positions of the putative MyoD binding sites are also indicated. A further enlargement indicates the location of the three fragments (F1, F2 and F3) amplified in the ChIP assay shown in the bottom. Bottom: In vivo binding activity of MyoD during differentiation. Chromatin from C2.7 myoblasts and MyoD-expressing fibroblasts (responsive and unresponsive) kept either in growth (T0) or in differentiation medium for 24 h (T24) was immunoprecipitated using a specific antibody to MyoD or in the absence of antibody (NoAb). Input represents non-immunoprecipitated cross-linked chromatin. The indicated fragments were amplified by PCR as described in ‘Materials and Methods’ section. A p57 promoter fragment, which we have previously demonstrated not to be bound by MyoD (28), was used as a negative control. The results obtained with 30 amplification cycles are shown. (b) qRT-PCR analysis and quantitation of the ChIP assay for MyoD binding to fragment 3 (F3) and myogenin (myog) promoter. Values derived from three independent experiments were normalized for background signals (NoAb) and expressed as percentage of input chromatin (% input). Data are shown as mean ± standard deviation.