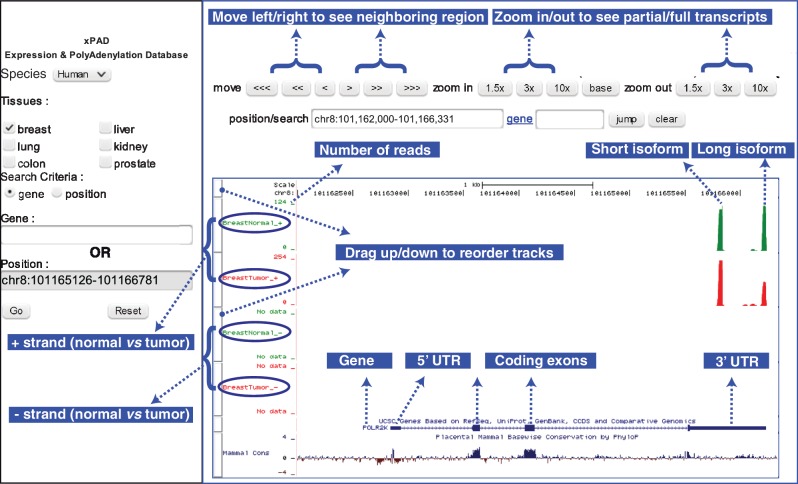
Figure 5.
Illustration of xPAD. xPAD integrates the UCSC genome browser to provide a web-interface to visualize both the precise polyadenylation locations of different isoforms, as well as their expression levels across tissues of interest. The complete gene structure of POLR2K highlights the utility of DRS; in both normal and tumor tissues, all polyadenylation sites exclusively occur in the 3′ UTR of the gene and within the sense strand, and the 3′ end of the reads (green/red bars) mapping to the long isoform matches within 2 nts of the 3′ UTR polyadenylation site. Normal breast contains 120 reads of long, and 109 reads of short isoforms, whereas breast tumor contains 257 reads of short isoform, which is upregulated, and 134 reads of long isoform that is almost unchanged. For brevity, many additional features such as evolutionary conservation (bottom track) and methylation marks (not shown), which are available via the UCSC browser panel (right) are not illustrated.