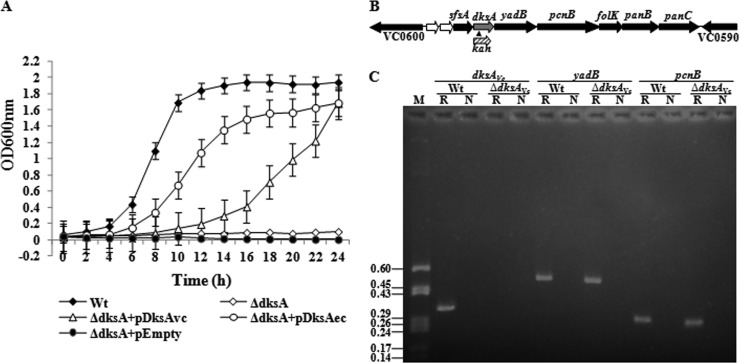
Fig 1.
(A) Growth complementation of the ΔdksAEc strain with a functional dksAVc gene in M9M medium. E. coli strains used are as follows: Wt, CF1648; ΔdksA, CF9240; ΔdksA+pDksAvc, CF9240(pDDKW1); ΔdksA+pDksAec, CF9240(pJK537); ΔdksA+pEmpty, CF9240(pDrive). Error bars indicate standard deviations. (B) Genomic arrangement of the dksA gene (gray arrow), including its flanking genetic determinants (VC0590 to VC0600), in V. cholerae. The direction of each arrow indicates the direction of transcription of a gene. VC0598 and VC0599 are two small hypothetical ORFs (white arrows). The insertion location of the kanamycin resistance gene (kan) cassette (small filled triangle) and its direction of transcription are also shown. (C) RT-PCR analysis to show that the deletion of dksAVc did not hamper transcription of downstream genes. V. cholerae strains used are the Wt (C6709) and the ΔdksAVc mutant (C-DksA1). Lanes: M, pBluescript II KS(+) plasmid DNA digested with HaeIII, used as markers; sizes (in kb) of the DNA fragments are given in the left margin; R, RT with Taq DNA polymerase; N, only Taq DNA polymerase (used as a negative control).