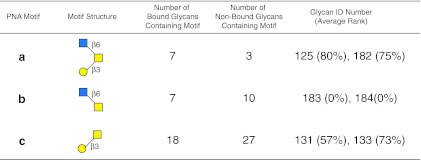
FIG. 7.
Display of motifs for PNA. The structures of motifs (a–c) discovered for PNA over three concentrations are indicated using symbols defined in Figure 1, with α and β anomeric carbons and linkage positions to the adjacent monosaccharides indicated. The glycan ID number indicating the position on v4.0 of the CFG glycan microarray (Supplementary Table S3; see online supplementary material at http://www.liebertpub.com) is shown for the motif found as a glycan on the array with its corresponding average rank in parentheses. Motifs discovered by the algorithm that are not found as glycans on the array have no ID number or ranking and are designated NA (not applicable). The number of bound glycans containing motif is determined by the algorithm, and indicates the number of bound glycans found on the glycan array that contain the corresponding motif, while the number of non-bound glycans containing motif indicates the number of glycans found on the glycan array that contain the motif, but are considered non-binding glycans by the algorithm. The display of the graphical user interface used to generate this summary is shown in Supplementary Figure S4; see online supplementary material at http://www.liebertpub.com).