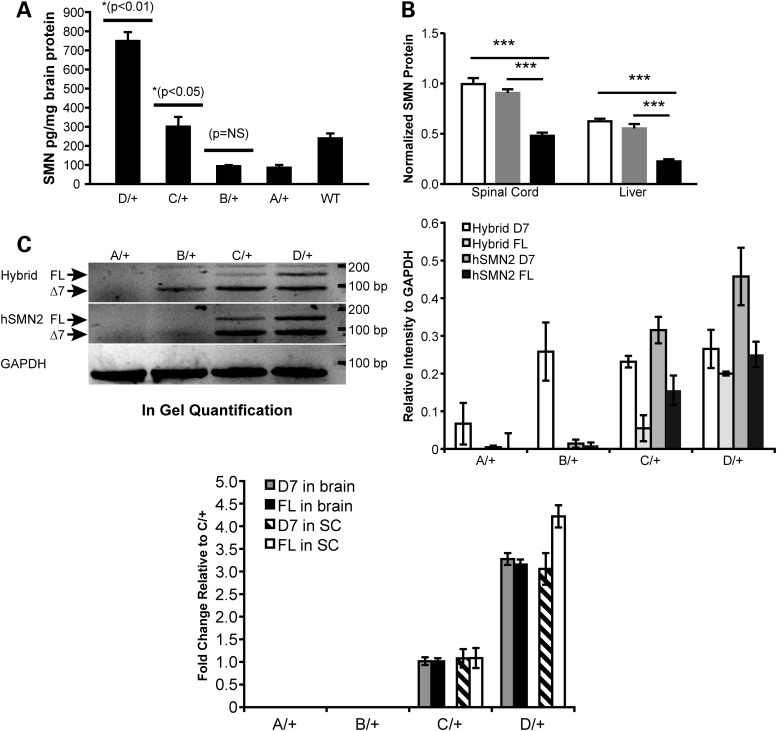
Figure 2.
SMN expression in allelic series mutants. (A) Total SMN protein detected by ELISA in brain lysates (n = 2/genotype) of heterozygous Smn1D, Smn1C, Smn1B, Smn1A and Smn1+/+ animals demonstrated a titration correlating with total numbers of relative SMN copies per allele. Heterozygous Smn1C allele mice have significantly more SMN in the brain than both heterozygous Smn1B and heterozygous Smn1A (P < 0.05; t-test), and significantly less SMN in the brain than heterozygous Smn1D allele mice (P < 0.01; t-test). Smn1+/+ protein levels demonstrate a preferential detection of human SMN relative to murine SMN protein. (B) Western blot analysis of FL SMN in the spinal cord and liver extracts of homozygous Smn1C/Cmutants demonstrated reduced relative SMN total protein (mouse + human) compared with heterozygous and WT controls. SMN protein levels were significantly (P < 0.001) reduced in the spinal cord and liver of homozygous Smn1C/C mutants (black bars). No differences were observed between heterozygous (gray bars) and WT controls (open bars). Data are represented as the mean ± SEM (n = 12 animals/genotype mixed genders) normalized first to ATP5B protein, then to WT levels. (C) Semi-qPCR of hybrid and human SMN. FL-SMN and D7-SMN were visualized based on the difference of product size on ethidium bromide-stained agarose gel. Left panel: FL, 148 bp, delta7: 94 bp. Right panel: Quantification of different products normalized to Gapdh on three animals/genotype. (D) Real-time RT-PCR analysis of D7-SMN2 versus FL-SMN2 derived from human genomic portions across the allelic series in total RNA isolated from the brain and the spinal cord. Data are represented as fold change values normalized to Smn1C/+.