Figure 3.
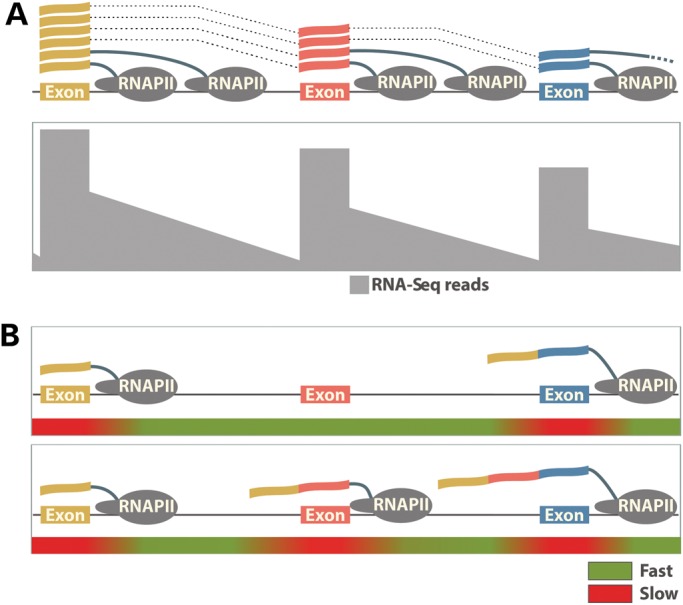
Co-transcriptional splicing and kinetic control of alternative splicing by the elongating RNAPII. (A) The genomic region of a hypothetical gene is illustrated containing three exons. Along the gene body, the frequency of RNA-Seq reads from a cell type where total RNA has been harvested is depicted. A characteristic ‘saw-tooth’ pattern is formed by the frequency of RNA-Seq reads across the genomic region. Exonic reads are most prevalent due to the amount of both nascent and partially processed RNAs containing these sequences. Intronic reads are also present albeit at lower levels representing unprocessed RNA species. The diminished frequency of reads toward the 3′ end of introns arises from rapid co-transcriptional splicing that occurs in this region after RNAPII finishes transcribing each intron. (B) The RNAPII elongation rate controls the exon inclusion. Rapid elongation through an alternative exon defined by weak splicing signals does not allow enough time for exon recognition before the competing strong splicing signals of the downstream exon are presented (top). As a result, the alternative exon is skipped. Slowing down RNAPII within and downstream of the alternative exon provides time for the splicing machinery to recognize and splice the weak exon in the mature transcript (bottom).
